| schema location: | ..\tcs_v101.xsd |
| attribute form default: | unqualified |
| element form default: | qualified |
| targetNamespace: | http://www.tdwg.org/schemas/tcs/1.01 |
element DataSet
| diagram | 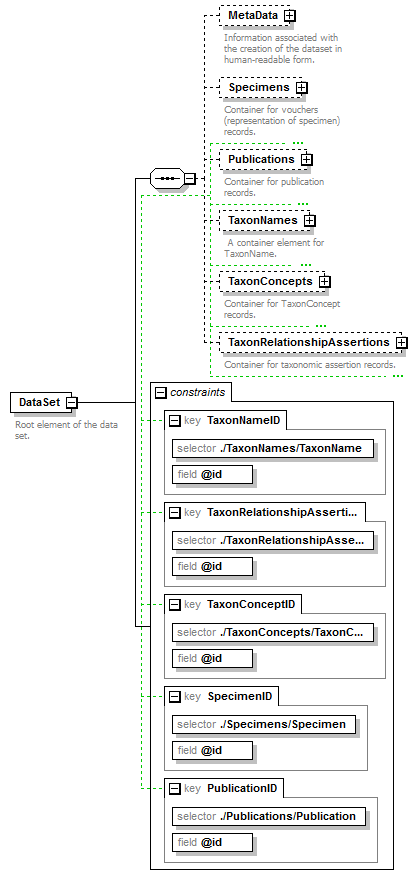 |
||||||||||||||||||||||||||||||
| namespace | http://www.tdwg.org/schemas/tcs/1.01 | ||||||||||||||||||||||||||||||
| properties |
|
||||||||||||||||||||||||||||||
| children | MetaData Specimens Publications TaxonNames TaxonConcepts TaxonRelationshipAssertions | ||||||||||||||||||||||||||||||
| identity constraints |
|
||||||||||||||||||||||||||||||
| annotation |
|
||||||||||||||||||||||||||||||
| source | <xs:element name="DataSet"> <xs:annotation> <xs:documentation>Root element of the data set.</xs:documentation> </xs:annotation> <xs:complexType> <xs:sequence> <xs:element name="MetaData" minOccurs="0"> <xs:annotation> <xs:documentation>Information associated with the creation of the dataset in human-readable form.</xs:documentation> </xs:annotation> <xs:complexType> <xs:sequence> <xs:element name="Simple" type="xs:string"> <xs:annotation> <xs:documentation>A summary of the metadata for this document.</xs:documentation> </xs:annotation> </xs:element> <xs:element name="MetaDataDetailed" type="PlaceholderType" minOccurs="0"> <xs:annotation> <xs:documentation>A place holder for an external schema giving details of the metadata for this document. A meta data schema is provided with the TCS release for this purpose should no other schema be preferred. </xs:documentation> </xs:annotation> </xs:element> </xs:sequence> </xs:complexType> </xs:element> <xs:element name="Specimens" minOccurs="0"> <xs:annotation> <xs:documentation>Container for vouchers (representation of specimen) records.</xs:documentation> </xs:annotation> <xs:complexType> <xs:sequence> <xs:element name="Specimen" minOccurs="0" maxOccurs="unbounded"> <xs:annotation> <xs:documentation>Specimen and location</xs:documentation> </xs:annotation> <xs:complexType> <xs:sequence minOccurs="0"> <xs:element name="Simple" type="xs:string"/> <xs:element name="Institution" minOccurs="0"> <xs:annotation> <xs:documentation>Information about the location of the specimen. The PlaceholderType represents a placeholder for optionally extending the schema with a schema element defined in another standard. </xs:documentation> </xs:annotation> <xs:complexType> <xs:sequence minOccurs="0"> <xs:element name="InstitutionName" type="xs:string"/> <xs:element name="Code" type="xs:string" minOccurs="0"/> <xs:element name="Address" type="xs:string" minOccurs="0"/> <xs:element name="Phone" type="xs:string" minOccurs="0"/> <xs:element name="URL" type="xs:string" minOccurs="0"/> <xs:element name="Email" type="xs:string" minOccurs="0"/> </xs:sequence> <xs:attribute name="identifier" type="xs:token" use="required"/> </xs:complexType> </xs:element> <xs:element name="Collection" minOccurs="0"> <xs:annotation> <xs:documentation>The PlaceholderType represents a placeholder for optionally extending the schema with a schema element defined in another standard.</xs:documentation> </xs:annotation> <xs:complexType> <xs:complexContent> <xs:extension base="PlaceholderType"> <xs:attribute name="identifier" type="xs:token" use="required"> <xs:annotation> <xs:documentation> Identifier for the specimen used within the collection. </xs:documentation> </xs:annotation> </xs:attribute> </xs:extension> </xs:complexContent> </xs:complexType> </xs:element> <xs:element name="SpecimenItem"> <xs:annotation> <xs:documentation>Physical object the voucher is referring to. The PlaceholderType represents a placeholder for optionally extending the schema with a schema element defined in another standard.</xs:documentation> </xs:annotation> <xs:complexType> <xs:complexContent> <xs:extension base="PlaceholderType"> <xs:attribute name="identifier" type="xs:token" use="required"/> </xs:extension> </xs:complexContent> </xs:complexType> </xs:element> </xs:sequence> <xs:attribute name="id" type="xs:token" use="required"> <xs:annotation> <xs:documentation>Unique identifier (key) of the element. It can be local to the data set or a GUID in which case it can be resolved to a location on the internet where the record is stored. </xs:documentation> </xs:annotation> </xs:attribute> </xs:complexType> </xs:element> </xs:sequence> </xs:complexType> </xs:element> <xs:element name="Publications" minOccurs="0"> <xs:annotation> <xs:documentation>Container for publication records.</xs:documentation> </xs:annotation> <xs:complexType> <xs:sequence> <xs:element name="Publication" minOccurs="0" maxOccurs="unbounded"> <xs:annotation> <xs:documentation>Details of the data source.</xs:documentation> </xs:annotation> <xs:complexType> <xs:sequence minOccurs="0"> <xs:element name="Simple" type="xs:string"> <xs:annotation> <xs:documentation>Full bibliographic reference as a single formatted string.</xs:documentation> </xs:annotation> </xs:element> <xs:element name="PublicationDetailed" type="PlaceholderType" minOccurs="0"> <xs:annotation> <xs:documentation>Mechanism to allow for the extension of the schema for specific applications.</xs:documentation> </xs:annotation> </xs:element> </xs:sequence> <xs:attribute name="id" type="xs:token" use="required"> <xs:annotation> <xs:documentation>Unique identifier (key) of the element. It can be local to the data set or a GUID in which case it can be resolved to a location on the internet where the record is stored. </xs:documentation> </xs:annotation> </xs:attribute> </xs:complexType> </xs:element> </xs:sequence> </xs:complexType> </xs:element> <xs:element name="TaxonNames" minOccurs="0"> <xs:annotation> <xs:documentation> A container element for TaxonName. </xs:documentation> </xs:annotation> <xs:complexType> <xs:sequence> <xs:element name="TaxonName" type="ScientificName" minOccurs="0" maxOccurs="unbounded"> <xs:annotation> <xs:documentation> An object that represents a single scientific biological name that either is, or appears to be, governed by one of the biological codes of nomenclature. These are not taxa. Taxa, whether accepted or not, are represented by TaxonConcept objects. Vernacular names are also dealt with under taxon concepts. </xs:documentation> </xs:annotation> </xs:element> </xs:sequence> </xs:complexType> </xs:element> <xs:element name="TaxonConcepts" minOccurs="0"> <xs:annotation> <xs:documentation>Container for TaxonConcept records.</xs:documentation> </xs:annotation> <xs:complexType> <xs:sequence> <xs:element name="TaxonConcept" minOccurs="0" maxOccurs="unbounded"> <xs:annotation> <xs:documentation> Representation of a TaxonConcept. Various types of concept can be represented, including a reference to the GUID of an existing Concept. </xs:documentation> </xs:annotation> <xs:complexType mixed="false"> <xs:sequence minOccurs="0"> <xs:element name="Name"> <xs:annotation> <xs:documentation>A non-unique handle to the concept. This can represent the name as published.</xs:documentation> </xs:annotation> <xs:complexType> <xs:complexContent> <xs:extension base="ReferenceType"> <xs:attribute name="scientific" type="xs:boolean" use="required"/> <xs:attribute name="language" type="xs:language" use="optional"/> </xs:extension> </xs:complexContent> </xs:complexType> </xs:element> <xs:element name="Rank" type="TaxonomicRank" minOccurs="0"> <xs:annotation> <xs:documentation> The taxonomic rank of this concept. Either as a string or as a code for a recognise rank or both. </xs:documentation> </xs:annotation> </xs:element> <xs:element name="AccordingTo" type="AccordingToType" minOccurs="0"> <xs:annotation> <xs:documentation>Information about the authorship of this concept which uses the Name in their sense (i.e. secundum, sensu).</xs:documentation> </xs:annotation> </xs:element> <xs:element name="TaxonRelationships" minOccurs="0"> <xs:annotation> <xs:documentation>Stores explicit, taxonomic and nomenclatural relationships that are part of the original concept definition.</xs:documentation> </xs:annotation> <xs:complexType> <xs:sequence> <xs:element name="TaxonRelationship" maxOccurs="unbounded"> <xs:annotation> <xs:documentation>Type of the relationship </xs:documentation> </xs:annotation> <xs:complexType> <xs:complexContent> <xs:extension base="RelationshipType"> <xs:sequence> <xs:element name="ToTaxonConcept" type="ReferenceType"> <xs:annotation> <xs:documentation>Reference to another TaxonConcept. </xs:documentation> </xs:annotation> </xs:element> </xs:sequence> </xs:extension> </xs:complexContent> </xs:complexType> </xs:element> </xs:sequence> </xs:complexType> </xs:element> <xs:element name="SpecimenCircumscription" minOccurs="0"> <xs:annotation> <xs:documentation>A set of specimens that are used to define the concept.</xs:documentation> </xs:annotation> <xs:complexType> <xs:sequence> <xs:element name="CircumscribedBy" type="ReferenceType" maxOccurs="unbounded"> <xs:annotation> <xs:documentation>Link to record of a specimen. </xs:documentation> </xs:annotation> </xs:element> </xs:sequence> </xs:complexType> </xs:element> <xs:element name="CharacterCircumscription" type="PlaceholderType" minOccurs="0"> <xs:annotation> <xs:documentation>A set of taxonomic descriptions used to define this concept. May potentially hold descriptions according to the TDWG SDD schema, or any other, format.</xs:documentation> </xs:annotation> </xs:element> <xs:element name="ProviderLink" type="xs:string" minOccurs="0"/> <xs:element name="ProviderSpecificData" type="PlaceholderType" minOccurs="0"> <xs:annotation> <xs:documentation>Mechanism to allow for the extension of the schema for specific applications.</xs:documentation> </xs:annotation> </xs:element> </xs:sequence> <xs:attribute name="id" type="xs:token" use="required"> <xs:annotation> <xs:documentation>Unique identifier (key) of the element. It can be local to the data set or a GUID in which case it can be resolved to a location on the internet where the record is stored. </xs:documentation> </xs:annotation> </xs:attribute> <xs:attribute name="type" use="optional"> <xs:annotation> <xs:documentation>The optional enumerated type of the Concept may reflect which data elements are provided.</xs:documentation> </xs:annotation> <xs:simpleType> <xs:restriction base="xs:string"> <xs:enumeration value="original"/> <xs:enumeration value="revision"/> <xs:enumeration value="incomplete"/> <xs:enumeration value="aggregate"/> <xs:enumeration value="nominal"/> </xs:restriction> </xs:simpleType> </xs:attribute> <xs:attribute name="primary" type="xs:boolean" use="optional"> <xs:annotation> <xs:documentation>If primary='true' the concept is the first level response to a query. If 'false' the concept may be a secondary concept linked directly or indirectly to the definition of a primary concept.</xs:documentation> </xs:annotation> </xs:attribute> <xs:attribute name="form" use="optional"> <xs:annotation> <xs:documentation> Flag as to sexual form or parentage. List of options may extend in future but are mutually exclusive. </xs:documentation> </xs:annotation> <xs:simpleType> <xs:restriction base="xs:string"> <xs:enumeration value="anamorph"/> <xs:enumeration value="teleomorph"/> <xs:enumeration value="hybrid"/> </xs:restriction> </xs:simpleType> </xs:attribute> </xs:complexType> </xs:element> </xs:sequence> </xs:complexType> </xs:element> <xs:element name="TaxonRelationshipAssertions" minOccurs="0"> <xs:annotation> <xs:documentation>Container for taxonomic assertion records.</xs:documentation> </xs:annotation> <xs:complexType> <xs:sequence> <xs:element name="TaxonRelationshipAssertion" minOccurs="0" maxOccurs="unbounded"> <xs:annotation> <xs:documentation>Relationships between two concepts which are not part of the original definition of either of these concepts; possibly by a third party. </xs:documentation> </xs:annotation> <xs:complexType> <xs:complexContent> <xs:extension base="RelationshipType"> <xs:sequence minOccurs="0"> <xs:element name="AccordingTo" type="AccordingToType"> <xs:annotation> <xs:documentation>Information about the authorship of the asserted relationship.</xs:documentation> </xs:annotation> </xs:element> <xs:element name="FromTaxonConcept"> <xs:annotation> <xs:documentation>Starting point of the directed relationship. A reference to a TaxonConcept.</xs:documentation> </xs:annotation> <xs:complexType> <xs:complexContent> <xs:extension base="ReferenceType"/> </xs:complexContent> </xs:complexType> </xs:element> <xs:element name="ToTaxonConcept"> <xs:annotation> <xs:documentation>End point of the directed relationship. A reference to a TaxonConcept.</xs:documentation> </xs:annotation> <xs:complexType> <xs:complexContent> <xs:extension base="ReferenceType"/> </xs:complexContent> </xs:complexType> </xs:element> </xs:sequence> <xs:attribute name="id" type="xs:token" use="required"> <xs:annotation> <xs:documentation>Unique identifier (key) of the element. It can be local to the data set or a GUID in which case it can be resolved to a location on the internet where the record is stored. </xs:documentation> </xs:annotation> </xs:attribute> </xs:extension> </xs:complexContent> </xs:complexType> </xs:element> </xs:sequence> </xs:complexType> </xs:element> </xs:sequence> </xs:complexType> <xs:key name="TaxonNameID"> <xs:selector xpath="./TaxonNames/TaxonName"/> <xs:field xpath="@id"/> </xs:key> <xs:key name="TaxonRelationshipAssertionID"> <xs:selector xpath="./TaxonRelationshipAssertions/TaxonRelationshipAssertion"/> <xs:field xpath="@id"/> </xs:key> <xs:key name="TaxonConceptID"> <xs:selector xpath="./TaxonConcepts/TaxonConcept"/> <xs:field xpath="@id"/> </xs:key> <xs:key name="SpecimenID"> <xs:selector xpath="./Specimens/Specimen"/> <xs:field xpath="@id"/> </xs:key> <xs:key name="PublicationID"> <xs:selector xpath="./Publications/Publication"/> <xs:field xpath="@id"/> </xs:key> </xs:element> |
element DataSet/MetaData
| diagram | 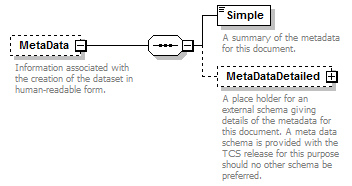 |
||||||||
| namespace | http://www.tdwg.org/schemas/tcs/1.01 | ||||||||
| properties |
|
||||||||
| children | Simple MetaDataDetailed | ||||||||
| annotation |
|
||||||||
| source | <xs:element name="MetaData" minOccurs="0"> <xs:annotation> <xs:documentation>Information associated with the creation of the dataset in human-readable form.</xs:documentation> </xs:annotation> <xs:complexType> <xs:sequence> <xs:element name="Simple" type="xs:string"> <xs:annotation> <xs:documentation>A summary of the metadata for this document.</xs:documentation> </xs:annotation> </xs:element> <xs:element name="MetaDataDetailed" type="PlaceholderType" minOccurs="0"> <xs:annotation> <xs:documentation>A place holder for an external schema giving details of the metadata for this document. A meta data schema is provided with the TCS release for this purpose should no other schema be preferred. </xs:documentation> </xs:annotation> </xs:element> </xs:sequence> </xs:complexType> </xs:element> |
element DataSet/MetaData/Simple
| diagram |  |
||||
| namespace | http://www.tdwg.org/schemas/tcs/1.01 | ||||
| type | xs:string | ||||
| properties |
|
||||
| annotation |
|
||||
| source | <xs:element name="Simple" type="xs:string"> <xs:annotation> <xs:documentation>A summary of the metadata for this document.</xs:documentation> </xs:annotation> </xs:element> |
element DataSet/MetaData/MetaDataDetailed
| diagram | 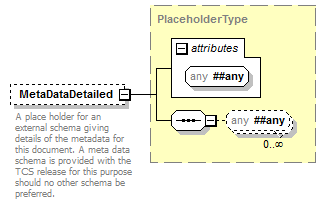 |
||||||||
| namespace | http://www.tdwg.org/schemas/tcs/1.01 | ||||||||
| type | PlaceholderType | ||||||||
| properties |
|
||||||||
| attributes |
|
||||||||
| annotation |
|
||||||||
| source | <xs:element name="MetaDataDetailed" type="PlaceholderType" minOccurs="0"> <xs:annotation> <xs:documentation>A place holder for an external schema giving details of the metadata for this document. A meta data schema is provided with the TCS release for this purpose should no other schema be preferred. </xs:documentation> </xs:annotation> </xs:element> |
element DataSet/Specimens
| diagram | 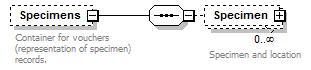 |
||||||||
| namespace | http://www.tdwg.org/schemas/tcs/1.01 | ||||||||
| properties |
|
||||||||
| children | Specimen | ||||||||
| annotation |
|
||||||||
| source | <xs:element name="Specimens" minOccurs="0"> <xs:annotation> <xs:documentation>Container for vouchers (representation of specimen) records.</xs:documentation> </xs:annotation> <xs:complexType> <xs:sequence> <xs:element name="Specimen" minOccurs="0" maxOccurs="unbounded"> <xs:annotation> <xs:documentation>Specimen and location</xs:documentation> </xs:annotation> <xs:complexType> <xs:sequence minOccurs="0"> <xs:element name="Simple" type="xs:string"/> <xs:element name="Institution" minOccurs="0"> <xs:annotation> <xs:documentation>Information about the location of the specimen. The PlaceholderType represents a placeholder for optionally extending the schema with a schema element defined in another standard. </xs:documentation> </xs:annotation> <xs:complexType> <xs:sequence minOccurs="0"> <xs:element name="InstitutionName" type="xs:string"/> <xs:element name="Code" type="xs:string" minOccurs="0"/> <xs:element name="Address" type="xs:string" minOccurs="0"/> <xs:element name="Phone" type="xs:string" minOccurs="0"/> <xs:element name="URL" type="xs:string" minOccurs="0"/> <xs:element name="Email" type="xs:string" minOccurs="0"/> </xs:sequence> <xs:attribute name="identifier" type="xs:token" use="required"/> </xs:complexType> </xs:element> <xs:element name="Collection" minOccurs="0"> <xs:annotation> <xs:documentation>The PlaceholderType represents a placeholder for optionally extending the schema with a schema element defined in another standard.</xs:documentation> </xs:annotation> <xs:complexType> <xs:complexContent> <xs:extension base="PlaceholderType"> <xs:attribute name="identifier" type="xs:token" use="required"> <xs:annotation> <xs:documentation> Identifier for the specimen used within the collection. </xs:documentation> </xs:annotation> </xs:attribute> </xs:extension> </xs:complexContent> </xs:complexType> </xs:element> <xs:element name="SpecimenItem"> <xs:annotation> <xs:documentation>Physical object the voucher is referring to. The PlaceholderType represents a placeholder for optionally extending the schema with a schema element defined in another standard.</xs:documentation> </xs:annotation> <xs:complexType> <xs:complexContent> <xs:extension base="PlaceholderType"> <xs:attribute name="identifier" type="xs:token" use="required"/> </xs:extension> </xs:complexContent> </xs:complexType> </xs:element> </xs:sequence> <xs:attribute name="id" type="xs:token" use="required"> <xs:annotation> <xs:documentation>Unique identifier (key) of the element. It can be local to the data set or a GUID in which case it can be resolved to a location on the internet where the record is stored. </xs:documentation> </xs:annotation> </xs:attribute> </xs:complexType> </xs:element> </xs:sequence> </xs:complexType> </xs:element> |
element DataSet/Specimens/Specimen
| diagram | 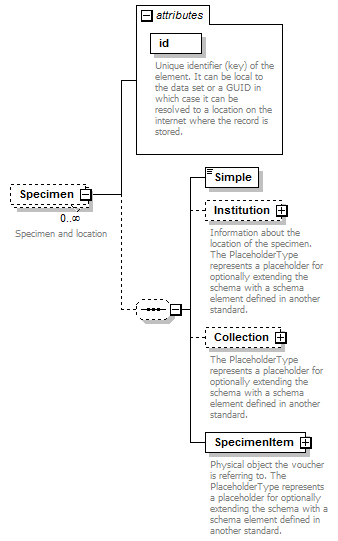 |
||||||||||||||
| namespace | http://www.tdwg.org/schemas/tcs/1.01 | ||||||||||||||
| properties |
|
||||||||||||||
| children | Simple Institution Collection SpecimenItem | ||||||||||||||
| attributes |
|
||||||||||||||
| annotation |
|
||||||||||||||
| source | <xs:element name="Specimen" minOccurs="0" maxOccurs="unbounded"> <xs:annotation> <xs:documentation>Specimen and location</xs:documentation> </xs:annotation> <xs:complexType> <xs:sequence minOccurs="0"> <xs:element name="Simple" type="xs:string"/> <xs:element name="Institution" minOccurs="0"> <xs:annotation> <xs:documentation>Information about the location of the specimen. The PlaceholderType represents a placeholder for optionally extending the schema with a schema element defined in another standard. </xs:documentation> </xs:annotation> <xs:complexType> <xs:sequence minOccurs="0"> <xs:element name="InstitutionName" type="xs:string"/> <xs:element name="Code" type="xs:string" minOccurs="0"/> <xs:element name="Address" type="xs:string" minOccurs="0"/> <xs:element name="Phone" type="xs:string" minOccurs="0"/> <xs:element name="URL" type="xs:string" minOccurs="0"/> <xs:element name="Email" type="xs:string" minOccurs="0"/> </xs:sequence> <xs:attribute name="identifier" type="xs:token" use="required"/> </xs:complexType> </xs:element> <xs:element name="Collection" minOccurs="0"> <xs:annotation> <xs:documentation>The PlaceholderType represents a placeholder for optionally extending the schema with a schema element defined in another standard.</xs:documentation> </xs:annotation> <xs:complexType> <xs:complexContent> <xs:extension base="PlaceholderType"> <xs:attribute name="identifier" type="xs:token" use="required"> <xs:annotation> <xs:documentation> Identifier for the specimen used within the collection. </xs:documentation> </xs:annotation> </xs:attribute> </xs:extension> </xs:complexContent> </xs:complexType> </xs:element> <xs:element name="SpecimenItem"> <xs:annotation> <xs:documentation>Physical object the voucher is referring to. The PlaceholderType represents a placeholder for optionally extending the schema with a schema element defined in another standard.</xs:documentation> </xs:annotation> <xs:complexType> <xs:complexContent> <xs:extension base="PlaceholderType"> <xs:attribute name="identifier" type="xs:token" use="required"/> </xs:extension> </xs:complexContent> </xs:complexType> </xs:element> </xs:sequence> <xs:attribute name="id" type="xs:token" use="required"> <xs:annotation> <xs:documentation>Unique identifier (key) of the element. It can be local to the data set or a GUID in which case it can be resolved to a location on the internet where the record is stored. </xs:documentation> </xs:annotation> </xs:attribute> </xs:complexType> </xs:element> |
attribute DataSet/Specimens/Specimen/@id
| type | xs:token | ||||
| properties |
|
||||
| annotation |
|
||||
| source | <xs:attribute name="id" type="xs:token" use="required"> <xs:annotation> <xs:documentation>Unique identifier (key) of the element. It can be local to the data set or a GUID in which case it can be resolved to a location on the internet where the record is stored. </xs:documentation> </xs:annotation> </xs:attribute> |
element DataSet/Specimens/Specimen/Simple
| diagram |  |
||||
| namespace | http://www.tdwg.org/schemas/tcs/1.01 | ||||
| type | xs:string | ||||
| properties |
|
||||
| source | <xs:element name="Simple" type="xs:string"/> |
element DataSet/Specimens/Specimen/Institution
| diagram | 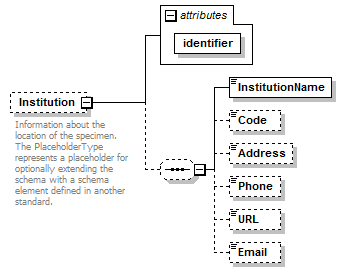 |
||||||||||||
| namespace | http://www.tdwg.org/schemas/tcs/1.01 | ||||||||||||
| properties |
|
||||||||||||
| children | InstitutionName Code Address Phone URL Email | ||||||||||||
| attributes |
|
||||||||||||
| annotation |
|
||||||||||||
| source | <xs:element name="Institution" minOccurs="0"> <xs:annotation> <xs:documentation>Information about the location of the specimen. The PlaceholderType represents a placeholder for optionally extending the schema with a schema element defined in another standard. </xs:documentation> </xs:annotation> <xs:complexType> <xs:sequence minOccurs="0"> <xs:element name="InstitutionName" type="xs:string"/> <xs:element name="Code" type="xs:string" minOccurs="0"/> <xs:element name="Address" type="xs:string" minOccurs="0"/> <xs:element name="Phone" type="xs:string" minOccurs="0"/> <xs:element name="URL" type="xs:string" minOccurs="0"/> <xs:element name="Email" type="xs:string" minOccurs="0"/> </xs:sequence> <xs:attribute name="identifier" type="xs:token" use="required"/> </xs:complexType> </xs:element> |
attribute DataSet/Specimens/Specimen/Institution/@identifier
| type | xs:token | ||||
| properties |
|
||||
| source | <xs:attribute name="identifier" type="xs:token" use="required"/> |
element DataSet/Specimens/Specimen/Institution/InstitutionName
| diagram |  |
||||
| namespace | http://www.tdwg.org/schemas/tcs/1.01 | ||||
| type | xs:string | ||||
| properties |
|
||||
| source | <xs:element name="InstitutionName" type="xs:string"/> |
element DataSet/Specimens/Specimen/Institution/Code
| diagram |  |
||||||||
| namespace | http://www.tdwg.org/schemas/tcs/1.01 | ||||||||
| type | xs:string | ||||||||
| properties |
|
||||||||
| source | <xs:element name="Code" type="xs:string" minOccurs="0"/> |
element DataSet/Specimens/Specimen/Institution/Address
| diagram |  |
||||||||
| namespace | http://www.tdwg.org/schemas/tcs/1.01 | ||||||||
| type | xs:string | ||||||||
| properties |
|
||||||||
| source | <xs:element name="Address" type="xs:string" minOccurs="0"/> |
element DataSet/Specimens/Specimen/Institution/Phone
| diagram |  |
||||||||
| namespace | http://www.tdwg.org/schemas/tcs/1.01 | ||||||||
| type | xs:string | ||||||||
| properties |
|
||||||||
| source | <xs:element name="Phone" type="xs:string" minOccurs="0"/> |
element DataSet/Specimens/Specimen/Institution/URL
| diagram |  |
||||||||
| namespace | http://www.tdwg.org/schemas/tcs/1.01 | ||||||||
| type | xs:string | ||||||||
| properties |
|
||||||||
| source | <xs:element name="URL" type="xs:string" minOccurs="0"/> |
element DataSet/Specimens/Specimen/Institution/Email
| diagram |  |
||||||||
| namespace | http://www.tdwg.org/schemas/tcs/1.01 | ||||||||
| type | xs:string | ||||||||
| properties |
|
||||||||
| source | <xs:element name="Email" type="xs:string" minOccurs="0"/> |
element DataSet/Specimens/Specimen/Collection
| diagram | 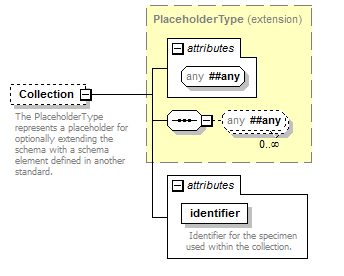 |
||||||||||||||
| namespace | http://www.tdwg.org/schemas/tcs/1.01 | ||||||||||||||
| type | extension of PlaceholderType | ||||||||||||||
| properties |
|
||||||||||||||
| attributes |
|
||||||||||||||
| annotation |
|
||||||||||||||
| source | <xs:element name="Collection" minOccurs="0"> <xs:annotation> <xs:documentation>The PlaceholderType represents a placeholder for optionally extending the schema with a schema element defined in another standard.</xs:documentation> </xs:annotation> <xs:complexType> <xs:complexContent> <xs:extension base="PlaceholderType"> <xs:attribute name="identifier" type="xs:token" use="required"> <xs:annotation> <xs:documentation> Identifier for the specimen used within the collection. </xs:documentation> </xs:annotation> </xs:attribute> </xs:extension> </xs:complexContent> </xs:complexType> </xs:element> |
attribute DataSet/Specimens/Specimen/Collection/@identifier
| type | xs:token | ||||
| properties |
|
||||
| annotation |
|
||||
| source | <xs:attribute name="identifier" type="xs:token" use="required"> <xs:annotation> <xs:documentation> Identifier for the specimen used within the collection. </xs:documentation> </xs:annotation> </xs:attribute> |
element DataSet/Specimens/Specimen/SpecimenItem
| diagram | 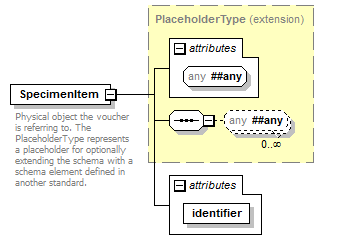 |
||||||||||||
| namespace | http://www.tdwg.org/schemas/tcs/1.01 | ||||||||||||
| type | extension of PlaceholderType | ||||||||||||
| properties |
|
||||||||||||
| attributes |
|
||||||||||||
| annotation |
|
||||||||||||
| source | <xs:element name="SpecimenItem"> <xs:annotation> <xs:documentation>Physical object the voucher is referring to. The PlaceholderType represents a placeholder for optionally extending the schema with a schema element defined in another standard.</xs:documentation> </xs:annotation> <xs:complexType> <xs:complexContent> <xs:extension base="PlaceholderType"> <xs:attribute name="identifier" type="xs:token" use="required"/> </xs:extension> </xs:complexContent> </xs:complexType> </xs:element> |
attribute DataSet/Specimens/Specimen/SpecimenItem/@identifier
| type | xs:token | ||||
| properties |
|
||||
| source | <xs:attribute name="identifier" type="xs:token" use="required"/> |
element DataSet/Publications
| diagram | 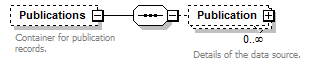 |
||||||||
| namespace | http://www.tdwg.org/schemas/tcs/1.01 | ||||||||
| properties |
|
||||||||
| children | Publication | ||||||||
| annotation |
|
||||||||
| source | <xs:element name="Publications" minOccurs="0"> <xs:annotation> <xs:documentation>Container for publication records.</xs:documentation> </xs:annotation> <xs:complexType> <xs:sequence> <xs:element name="Publication" minOccurs="0" maxOccurs="unbounded"> <xs:annotation> <xs:documentation>Details of the data source.</xs:documentation> </xs:annotation> <xs:complexType> <xs:sequence minOccurs="0"> <xs:element name="Simple" type="xs:string"> <xs:annotation> <xs:documentation>Full bibliographic reference as a single formatted string.</xs:documentation> </xs:annotation> </xs:element> <xs:element name="PublicationDetailed" type="PlaceholderType" minOccurs="0"> <xs:annotation> <xs:documentation>Mechanism to allow for the extension of the schema for specific applications.</xs:documentation> </xs:annotation> </xs:element> </xs:sequence> <xs:attribute name="id" type="xs:token" use="required"> <xs:annotation> <xs:documentation>Unique identifier (key) of the element. It can be local to the data set or a GUID in which case it can be resolved to a location on the internet where the record is stored. </xs:documentation> </xs:annotation> </xs:attribute> </xs:complexType> </xs:element> </xs:sequence> </xs:complexType> </xs:element> |
element DataSet/Publications/Publication
| diagram | 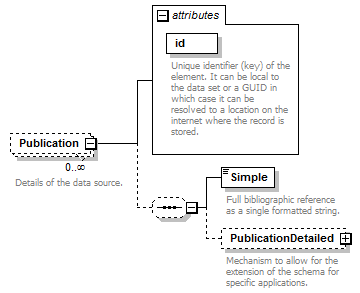 |
||||||||||||||
| namespace | http://www.tdwg.org/schemas/tcs/1.01 | ||||||||||||||
| properties |
|
||||||||||||||
| children | Simple PublicationDetailed | ||||||||||||||
| attributes |
|
||||||||||||||
| annotation |
|
||||||||||||||
| source | <xs:element name="Publication" minOccurs="0" maxOccurs="unbounded"> <xs:annotation> <xs:documentation>Details of the data source.</xs:documentation> </xs:annotation> <xs:complexType> <xs:sequence minOccurs="0"> <xs:element name="Simple" type="xs:string"> <xs:annotation> <xs:documentation>Full bibliographic reference as a single formatted string.</xs:documentation> </xs:annotation> </xs:element> <xs:element name="PublicationDetailed" type="PlaceholderType" minOccurs="0"> <xs:annotation> <xs:documentation>Mechanism to allow for the extension of the schema for specific applications.</xs:documentation> </xs:annotation> </xs:element> </xs:sequence> <xs:attribute name="id" type="xs:token" use="required"> <xs:annotation> <xs:documentation>Unique identifier (key) of the element. It can be local to the data set or a GUID in which case it can be resolved to a location on the internet where the record is stored. </xs:documentation> </xs:annotation> </xs:attribute> </xs:complexType> </xs:element> |
attribute DataSet/Publications/Publication/@id
| type | xs:token | ||||
| properties |
|
||||
| annotation |
|
||||
| source | <xs:attribute name="id" type="xs:token" use="required"> <xs:annotation> <xs:documentation>Unique identifier (key) of the element. It can be local to the data set or a GUID in which case it can be resolved to a location on the internet where the record is stored. </xs:documentation> </xs:annotation> </xs:attribute> |
element DataSet/Publications/Publication/Simple
| diagram |  |
||||
| namespace | http://www.tdwg.org/schemas/tcs/1.01 | ||||
| type | xs:string | ||||
| properties |
|
||||
| annotation |
|
||||
| source | <xs:element name="Simple" type="xs:string"> <xs:annotation> <xs:documentation>Full bibliographic reference as a single formatted string.</xs:documentation> </xs:annotation> </xs:element> |
element DataSet/Publications/Publication/PublicationDetailed
| diagram | 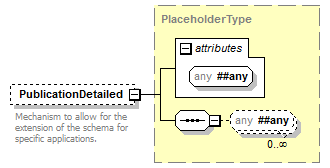 |
||||||||
| namespace | http://www.tdwg.org/schemas/tcs/1.01 | ||||||||
| type | PlaceholderType | ||||||||
| properties |
|
||||||||
| attributes |
|
||||||||
| annotation |
|
||||||||
| source | <xs:element name="PublicationDetailed" type="PlaceholderType" minOccurs="0"> <xs:annotation> <xs:documentation>Mechanism to allow for the extension of the schema for specific applications.</xs:documentation> </xs:annotation> </xs:element> |
element DataSet/TaxonNames
| diagram | 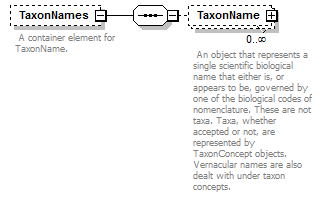 |
||||||||
| namespace | http://www.tdwg.org/schemas/tcs/1.01 | ||||||||
| properties |
|
||||||||
| children | TaxonName | ||||||||
| annotation |
|
||||||||
| source | <xs:element name="TaxonNames" minOccurs="0"> <xs:annotation> <xs:documentation> A container element for TaxonName. </xs:documentation> </xs:annotation> <xs:complexType> <xs:sequence> <xs:element name="TaxonName" type="ScientificName" minOccurs="0" maxOccurs="unbounded"> <xs:annotation> <xs:documentation> An object that represents a single scientific biological name that either is, or appears to be, governed by one of the biological codes of nomenclature. These are not taxa. Taxa, whether accepted or not, are represented by TaxonConcept objects. Vernacular names are also dealt with under taxon concepts. </xs:documentation> </xs:annotation> </xs:element> </xs:sequence> </xs:complexType> </xs:element> |
element DataSet/TaxonNames/TaxonName
| diagram | 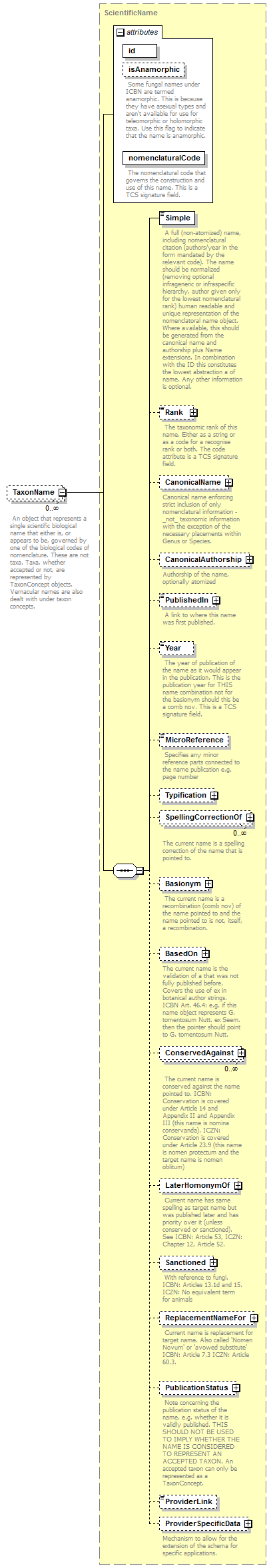 |
||||||||||||||||||||||||||||
| namespace | http://www.tdwg.org/schemas/tcs/1.01 | ||||||||||||||||||||||||||||
| type | ScientificName | ||||||||||||||||||||||||||||
| properties |
|
||||||||||||||||||||||||||||
| children | Simple Rank CanonicalName CanonicalAuthorship PublishedIn Year MicroReference Typification SpellingCorrectionOf Basionym BasedOn ConservedAgainst LaterHomonymOf Sanctioned ReplacementNameFor PublicationStatus ProviderLink ProviderSpecificData | ||||||||||||||||||||||||||||
| attributes |
|
||||||||||||||||||||||||||||
| annotation |
|
||||||||||||||||||||||||||||
| source | <xs:element name="TaxonName" type="ScientificName" minOccurs="0" maxOccurs="unbounded"> <xs:annotation> <xs:documentation> An object that represents a single scientific biological name that either is, or appears to be, governed by one of the biological codes of nomenclature. These are not taxa. Taxa, whether accepted or not, are represented by TaxonConcept objects. Vernacular names are also dealt with under taxon concepts. </xs:documentation> </xs:annotation> </xs:element> |
element DataSet/TaxonConcepts
| diagram | 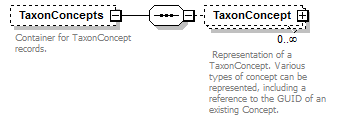 |
||||||||
| namespace | http://www.tdwg.org/schemas/tcs/1.01 | ||||||||
| properties |
|
||||||||
| children | TaxonConcept | ||||||||
| annotation |
|
||||||||
| source | <xs:element name="TaxonConcepts" minOccurs="0"> <xs:annotation> <xs:documentation>Container for TaxonConcept records.</xs:documentation> </xs:annotation> <xs:complexType> <xs:sequence> <xs:element name="TaxonConcept" minOccurs="0" maxOccurs="unbounded"> <xs:annotation> <xs:documentation> Representation of a TaxonConcept. Various types of concept can be represented, including a reference to the GUID of an existing Concept. </xs:documentation> </xs:annotation> <xs:complexType mixed="false"> <xs:sequence minOccurs="0"> <xs:element name="Name"> <xs:annotation> <xs:documentation>A non-unique handle to the concept. This can represent the name as published.</xs:documentation> </xs:annotation> <xs:complexType> <xs:complexContent> <xs:extension base="ReferenceType"> <xs:attribute name="scientific" type="xs:boolean" use="required"/> <xs:attribute name="language" type="xs:language" use="optional"/> </xs:extension> </xs:complexContent> </xs:complexType> </xs:element> <xs:element name="Rank" type="TaxonomicRank" minOccurs="0"> <xs:annotation> <xs:documentation> The taxonomic rank of this concept. Either as a string or as a code for a recognise rank or both. </xs:documentation> </xs:annotation> </xs:element> <xs:element name="AccordingTo" type="AccordingToType" minOccurs="0"> <xs:annotation> <xs:documentation>Information about the authorship of this concept which uses the Name in their sense (i.e. secundum, sensu).</xs:documentation> </xs:annotation> </xs:element> <xs:element name="TaxonRelationships" minOccurs="0"> <xs:annotation> <xs:documentation>Stores explicit, taxonomic and nomenclatural relationships that are part of the original concept definition.</xs:documentation> </xs:annotation> <xs:complexType> <xs:sequence> <xs:element name="TaxonRelationship" maxOccurs="unbounded"> <xs:annotation> <xs:documentation>Type of the relationship </xs:documentation> </xs:annotation> <xs:complexType> <xs:complexContent> <xs:extension base="RelationshipType"> <xs:sequence> <xs:element name="ToTaxonConcept" type="ReferenceType"> <xs:annotation> <xs:documentation>Reference to another TaxonConcept. </xs:documentation> </xs:annotation> </xs:element> </xs:sequence> </xs:extension> </xs:complexContent> </xs:complexType> </xs:element> </xs:sequence> </xs:complexType> </xs:element> <xs:element name="SpecimenCircumscription" minOccurs="0"> <xs:annotation> <xs:documentation>A set of specimens that are used to define the concept.</xs:documentation> </xs:annotation> <xs:complexType> <xs:sequence> <xs:element name="CircumscribedBy" type="ReferenceType" maxOccurs="unbounded"> <xs:annotation> <xs:documentation>Link to record of a specimen. </xs:documentation> </xs:annotation> </xs:element> </xs:sequence> </xs:complexType> </xs:element> <xs:element name="CharacterCircumscription" type="PlaceholderType" minOccurs="0"> <xs:annotation> <xs:documentation>A set of taxonomic descriptions used to define this concept. May potentially hold descriptions according to the TDWG SDD schema, or any other, format.</xs:documentation> </xs:annotation> </xs:element> <xs:element name="ProviderLink" type="xs:string" minOccurs="0"/> <xs:element name="ProviderSpecificData" type="PlaceholderType" minOccurs="0"> <xs:annotation> <xs:documentation>Mechanism to allow for the extension of the schema for specific applications.</xs:documentation> </xs:annotation> </xs:element> </xs:sequence> <xs:attribute name="id" type="xs:token" use="required"> <xs:annotation> <xs:documentation>Unique identifier (key) of the element. It can be local to the data set or a GUID in which case it can be resolved to a location on the internet where the record is stored. </xs:documentation> </xs:annotation> </xs:attribute> <xs:attribute name="type" use="optional"> <xs:annotation> <xs:documentation>The optional enumerated type of the Concept may reflect which data elements are provided.</xs:documentation> </xs:annotation> <xs:simpleType> <xs:restriction base="xs:string"> <xs:enumeration value="original"/> <xs:enumeration value="revision"/> <xs:enumeration value="incomplete"/> <xs:enumeration value="aggregate"/> <xs:enumeration value="nominal"/> </xs:restriction> </xs:simpleType> </xs:attribute> <xs:attribute name="primary" type="xs:boolean" use="optional"> <xs:annotation> <xs:documentation>If primary='true' the concept is the first level response to a query. If 'false' the concept may be a secondary concept linked directly or indirectly to the definition of a primary concept.</xs:documentation> </xs:annotation> </xs:attribute> <xs:attribute name="form" use="optional"> <xs:annotation> <xs:documentation> Flag as to sexual form or parentage. List of options may extend in future but are mutually exclusive. </xs:documentation> </xs:annotation> <xs:simpleType> <xs:restriction base="xs:string"> <xs:enumeration value="anamorph"/> <xs:enumeration value="teleomorph"/> <xs:enumeration value="hybrid"/> </xs:restriction> </xs:simpleType> </xs:attribute> </xs:complexType> </xs:element> </xs:sequence> </xs:complexType> </xs:element> |
element DataSet/TaxonConcepts/TaxonConcept
| diagram | 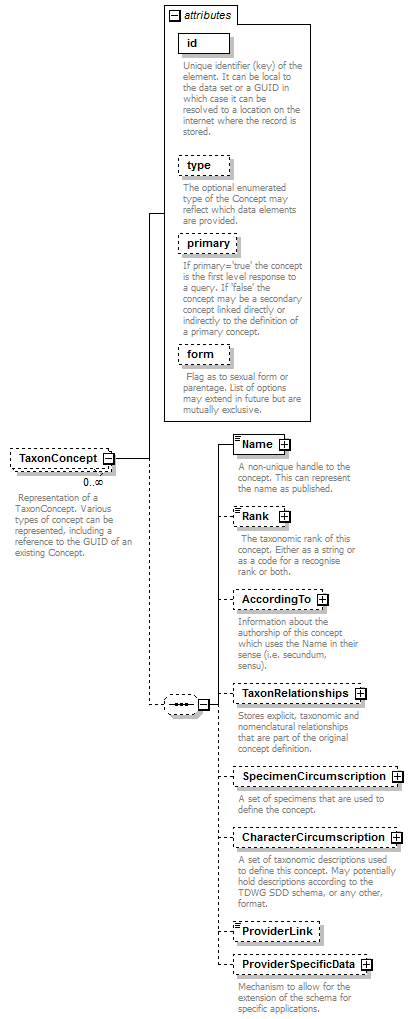 |
||||||||||||||||||||||||||||||||||||||
| namespace | http://www.tdwg.org/schemas/tcs/1.01 | ||||||||||||||||||||||||||||||||||||||
| properties |
|
||||||||||||||||||||||||||||||||||||||
| children | Name Rank AccordingTo TaxonRelationships SpecimenCircumscription CharacterCircumscription ProviderLink ProviderSpecificData | ||||||||||||||||||||||||||||||||||||||
| attributes |
|
||||||||||||||||||||||||||||||||||||||
| annotation |
|
||||||||||||||||||||||||||||||||||||||
| source | <xs:element name="TaxonConcept" minOccurs="0" maxOccurs="unbounded"> <xs:annotation> <xs:documentation> Representation of a TaxonConcept. Various types of concept can be represented, including a reference to the GUID of an existing Concept. </xs:documentation> </xs:annotation> <xs:complexType mixed="false"> <xs:sequence minOccurs="0"> <xs:element name="Name"> <xs:annotation> <xs:documentation>A non-unique handle to the concept. This can represent the name as published.</xs:documentation> </xs:annotation> <xs:complexType> <xs:complexContent> <xs:extension base="ReferenceType"> <xs:attribute name="scientific" type="xs:boolean" use="required"/> <xs:attribute name="language" type="xs:language" use="optional"/> </xs:extension> </xs:complexContent> </xs:complexType> </xs:element> <xs:element name="Rank" type="TaxonomicRank" minOccurs="0"> <xs:annotation> <xs:documentation> The taxonomic rank of this concept. Either as a string or as a code for a recognise rank or both. </xs:documentation> </xs:annotation> </xs:element> <xs:element name="AccordingTo" type="AccordingToType" minOccurs="0"> <xs:annotation> <xs:documentation>Information about the authorship of this concept which uses the Name in their sense (i.e. secundum, sensu).</xs:documentation> </xs:annotation> </xs:element> <xs:element name="TaxonRelationships" minOccurs="0"> <xs:annotation> <xs:documentation>Stores explicit, taxonomic and nomenclatural relationships that are part of the original concept definition.</xs:documentation> </xs:annotation> <xs:complexType> <xs:sequence> <xs:element name="TaxonRelationship" maxOccurs="unbounded"> <xs:annotation> <xs:documentation>Type of the relationship </xs:documentation> </xs:annotation> <xs:complexType> <xs:complexContent> <xs:extension base="RelationshipType"> <xs:sequence> <xs:element name="ToTaxonConcept" type="ReferenceType"> <xs:annotation> <xs:documentation>Reference to another TaxonConcept. </xs:documentation> </xs:annotation> </xs:element> </xs:sequence> </xs:extension> </xs:complexContent> </xs:complexType> </xs:element> </xs:sequence> </xs:complexType> </xs:element> <xs:element name="SpecimenCircumscription" minOccurs="0"> <xs:annotation> <xs:documentation>A set of specimens that are used to define the concept.</xs:documentation> </xs:annotation> <xs:complexType> <xs:sequence> <xs:element name="CircumscribedBy" type="ReferenceType" maxOccurs="unbounded"> <xs:annotation> <xs:documentation>Link to record of a specimen. </xs:documentation> </xs:annotation> </xs:element> </xs:sequence> </xs:complexType> </xs:element> <xs:element name="CharacterCircumscription" type="PlaceholderType" minOccurs="0"> <xs:annotation> <xs:documentation>A set of taxonomic descriptions used to define this concept. May potentially hold descriptions according to the TDWG SDD schema, or any other, format.</xs:documentation> </xs:annotation> </xs:element> <xs:element name="ProviderLink" type="xs:string" minOccurs="0"/> <xs:element name="ProviderSpecificData" type="PlaceholderType" minOccurs="0"> <xs:annotation> <xs:documentation>Mechanism to allow for the extension of the schema for specific applications.</xs:documentation> </xs:annotation> </xs:element> </xs:sequence> <xs:attribute name="id" type="xs:token" use="required"> <xs:annotation> <xs:documentation>Unique identifier (key) of the element. It can be local to the data set or a GUID in which case it can be resolved to a location on the internet where the record is stored. </xs:documentation> </xs:annotation> </xs:attribute> <xs:attribute name="type" use="optional"> <xs:annotation> <xs:documentation>The optional enumerated type of the Concept may reflect which data elements are provided.</xs:documentation> </xs:annotation> <xs:simpleType> <xs:restriction base="xs:string"> <xs:enumeration value="original"/> <xs:enumeration value="revision"/> <xs:enumeration value="incomplete"/> <xs:enumeration value="aggregate"/> <xs:enumeration value="nominal"/> </xs:restriction> </xs:simpleType> </xs:attribute> <xs:attribute name="primary" type="xs:boolean" use="optional"> <xs:annotation> <xs:documentation>If primary='true' the concept is the first level response to a query. If 'false' the concept may be a secondary concept linked directly or indirectly to the definition of a primary concept.</xs:documentation> </xs:annotation> </xs:attribute> <xs:attribute name="form" use="optional"> <xs:annotation> <xs:documentation> Flag as to sexual form or parentage. List of options may extend in future but are mutually exclusive. </xs:documentation> </xs:annotation> <xs:simpleType> <xs:restriction base="xs:string"> <xs:enumeration value="anamorph"/> <xs:enumeration value="teleomorph"/> <xs:enumeration value="hybrid"/> </xs:restriction> </xs:simpleType> </xs:attribute> </xs:complexType> </xs:element> |
attribute DataSet/TaxonConcepts/TaxonConcept/@id
| type | xs:token | ||||
| properties |
|
||||
| annotation |
|
||||
| source | <xs:attribute name="id" type="xs:token" use="required"> <xs:annotation> <xs:documentation>Unique identifier (key) of the element. It can be local to the data set or a GUID in which case it can be resolved to a location on the internet where the record is stored. </xs:documentation> </xs:annotation> </xs:attribute> |
attribute DataSet/TaxonConcepts/TaxonConcept/@type
| type | restriction of xs:string | ||||||||||||||||||
| properties |
|
||||||||||||||||||
| facets |
|
||||||||||||||||||
| annotation |
|
||||||||||||||||||
| source | <xs:attribute name="type" use="optional"> <xs:annotation> <xs:documentation>The optional enumerated type of the Concept may reflect which data elements are provided.</xs:documentation> </xs:annotation> <xs:simpleType> <xs:restriction base="xs:string"> <xs:enumeration value="original"/> <xs:enumeration value="revision"/> <xs:enumeration value="incomplete"/> <xs:enumeration value="aggregate"/> <xs:enumeration value="nominal"/> </xs:restriction> </xs:simpleType> </xs:attribute> |
attribute DataSet/TaxonConcepts/TaxonConcept/@primary
| type | xs:boolean | ||||
| properties |
|
||||
| annotation |
|
||||
| source | <xs:attribute name="primary" type="xs:boolean" use="optional"> <xs:annotation> <xs:documentation>If primary='true' the concept is the first level response to a query. If 'false' the concept may be a secondary concept linked directly or indirectly to the definition of a primary concept.</xs:documentation> </xs:annotation> </xs:attribute> |
attribute DataSet/TaxonConcepts/TaxonConcept/@form
| type | restriction of xs:string | ||||||||||||
| properties |
|
||||||||||||
| facets |
|
||||||||||||
| annotation |
|
||||||||||||
| source | <xs:attribute name="form" use="optional"> <xs:annotation> <xs:documentation> Flag as to sexual form or parentage. List of options may extend in future but are mutually exclusive. </xs:documentation> </xs:annotation> <xs:simpleType> <xs:restriction base="xs:string"> <xs:enumeration value="anamorph"/> <xs:enumeration value="teleomorph"/> <xs:enumeration value="hybrid"/> </xs:restriction> </xs:simpleType> </xs:attribute> |
element DataSet/TaxonConcepts/TaxonConcept/Name
| diagram | 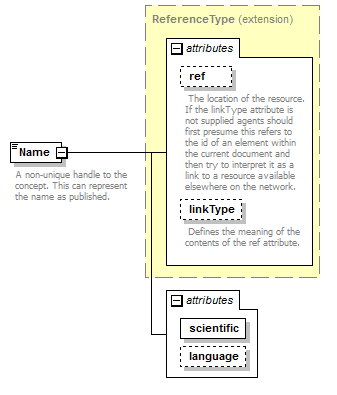 |
||||||||||||||||||||||||||||||||||
| namespace | http://www.tdwg.org/schemas/tcs/1.01 | ||||||||||||||||||||||||||||||||||
| type | extension of ReferenceType | ||||||||||||||||||||||||||||||||||
| properties |
|
||||||||||||||||||||||||||||||||||
| attributes |
|
||||||||||||||||||||||||||||||||||
| annotation |
|
||||||||||||||||||||||||||||||||||
| source | <xs:element name="Name"> <xs:annotation> <xs:documentation>A non-unique handle to the concept. This can represent the name as published.</xs:documentation> </xs:annotation> <xs:complexType> <xs:complexContent> <xs:extension base="ReferenceType"> <xs:attribute name="scientific" type="xs:boolean" use="required"/> <xs:attribute name="language" type="xs:language" use="optional"/> </xs:extension> </xs:complexContent> </xs:complexType> </xs:element> |
attribute DataSet/TaxonConcepts/TaxonConcept/Name/@scientific
| type | xs:boolean | ||||
| properties |
|
||||
| source | <xs:attribute name="scientific" type="xs:boolean" use="required"/> |
attribute DataSet/TaxonConcepts/TaxonConcept/Name/@language
| type | xs:language | ||||
| properties |
|
||||
| source | <xs:attribute name="language" type="xs:language" use="optional"/> |
element DataSet/TaxonConcepts/TaxonConcept/Rank
| diagram | 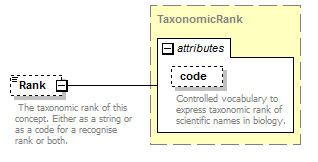 |
||||||||||||||
| namespace | http://www.tdwg.org/schemas/tcs/1.01 | ||||||||||||||
| type | TaxonomicRank | ||||||||||||||
| properties |
|
||||||||||||||
| attributes |
|
||||||||||||||
| annotation |
|
||||||||||||||
| source | <xs:element name="Rank" type="TaxonomicRank" minOccurs="0"> <xs:annotation> <xs:documentation> The taxonomic rank of this concept. Either as a string or as a code for a recognise rank or both. </xs:documentation> </xs:annotation> </xs:element> |
element DataSet/TaxonConcepts/TaxonConcept/AccordingTo
| diagram | 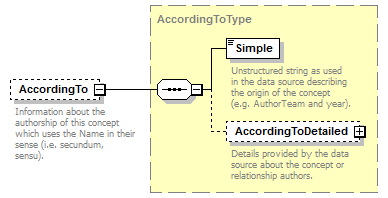 |
||||||||
| namespace | http://www.tdwg.org/schemas/tcs/1.01 | ||||||||
| type | AccordingToType | ||||||||
| properties |
|
||||||||
| children | Simple AccordingToDetailed | ||||||||
| annotation |
|
||||||||
| source | <xs:element name="AccordingTo" type="AccordingToType" minOccurs="0"> <xs:annotation> <xs:documentation>Information about the authorship of this concept which uses the Name in their sense (i.e. secundum, sensu).</xs:documentation> </xs:annotation> </xs:element> |
element DataSet/TaxonConcepts/TaxonConcept/TaxonRelationships
| diagram | 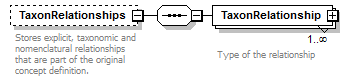 |
||||||||
| namespace | http://www.tdwg.org/schemas/tcs/1.01 | ||||||||
| properties |
|
||||||||
| children | TaxonRelationship | ||||||||
| annotation |
|
||||||||
| source | <xs:element name="TaxonRelationships" minOccurs="0"> <xs:annotation> <xs:documentation>Stores explicit, taxonomic and nomenclatural relationships that are part of the original concept definition.</xs:documentation> </xs:annotation> <xs:complexType> <xs:sequence> <xs:element name="TaxonRelationship" maxOccurs="unbounded"> <xs:annotation> <xs:documentation>Type of the relationship </xs:documentation> </xs:annotation> <xs:complexType> <xs:complexContent> <xs:extension base="RelationshipType"> <xs:sequence> <xs:element name="ToTaxonConcept" type="ReferenceType"> <xs:annotation> <xs:documentation>Reference to another TaxonConcept. </xs:documentation> </xs:annotation> </xs:element> </xs:sequence> </xs:extension> </xs:complexContent> </xs:complexType> </xs:element> </xs:sequence> </xs:complexType> </xs:element> |
element DataSet/TaxonConcepts/TaxonConcept/TaxonRelationships/TaxonRelationship
| diagram | 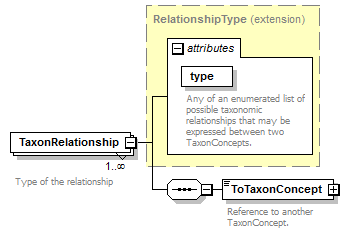 |
||||||||||||||
| namespace | http://www.tdwg.org/schemas/tcs/1.01 | ||||||||||||||
| type | extension of RelationshipType | ||||||||||||||
| properties |
|
||||||||||||||
| children | ToTaxonConcept | ||||||||||||||
| attributes |
|
||||||||||||||
| annotation |
|
||||||||||||||
| source | <xs:element name="TaxonRelationship" maxOccurs="unbounded"> <xs:annotation> <xs:documentation>Type of the relationship </xs:documentation> </xs:annotation> <xs:complexType> <xs:complexContent> <xs:extension base="RelationshipType"> <xs:sequence> <xs:element name="ToTaxonConcept" type="ReferenceType"> <xs:annotation> <xs:documentation>Reference to another TaxonConcept. </xs:documentation> </xs:annotation> </xs:element> </xs:sequence> </xs:extension> </xs:complexContent> </xs:complexType> </xs:element> |
element DataSet/TaxonConcepts/TaxonConcept/TaxonRelationships/TaxonRelationship/ToTaxonConcept
| diagram | 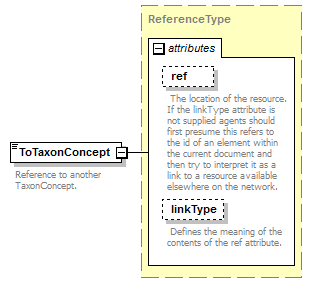 |
||||||||||||||||||||||
| namespace | http://www.tdwg.org/schemas/tcs/1.01 | ||||||||||||||||||||||
| type | ReferenceType | ||||||||||||||||||||||
| properties |
|
||||||||||||||||||||||
| attributes |
|
||||||||||||||||||||||
| annotation |
|
||||||||||||||||||||||
| source | <xs:element name="ToTaxonConcept" type="ReferenceType"> <xs:annotation> <xs:documentation>Reference to another TaxonConcept. </xs:documentation> </xs:annotation> </xs:element> |
element DataSet/TaxonConcepts/TaxonConcept/SpecimenCircumscription
| diagram | 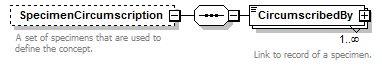 |
||||||||
| namespace | http://www.tdwg.org/schemas/tcs/1.01 | ||||||||
| properties |
|
||||||||
| children | CircumscribedBy | ||||||||
| annotation |
|
||||||||
| source | <xs:element name="SpecimenCircumscription" minOccurs="0"> <xs:annotation> <xs:documentation>A set of specimens that are used to define the concept.</xs:documentation> </xs:annotation> <xs:complexType> <xs:sequence> <xs:element name="CircumscribedBy" type="ReferenceType" maxOccurs="unbounded"> <xs:annotation> <xs:documentation>Link to record of a specimen. </xs:documentation> </xs:annotation> </xs:element> </xs:sequence> </xs:complexType> </xs:element> |
element DataSet/TaxonConcepts/TaxonConcept/SpecimenCircumscription/CircumscribedBy
| diagram | 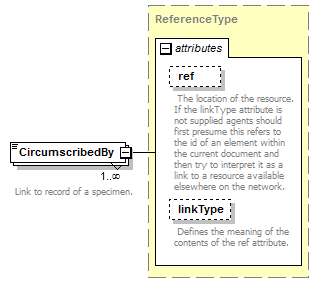 |
||||||||||||||||||||||
| namespace | http://www.tdwg.org/schemas/tcs/1.01 | ||||||||||||||||||||||
| type | ReferenceType | ||||||||||||||||||||||
| properties |
|
||||||||||||||||||||||
| attributes |
|
||||||||||||||||||||||
| annotation |
|
||||||||||||||||||||||
| source | <xs:element name="CircumscribedBy" type="ReferenceType" maxOccurs="unbounded"> <xs:annotation> <xs:documentation>Link to record of a specimen. </xs:documentation> </xs:annotation> </xs:element> |
element DataSet/TaxonConcepts/TaxonConcept/CharacterCircumscription
| diagram | 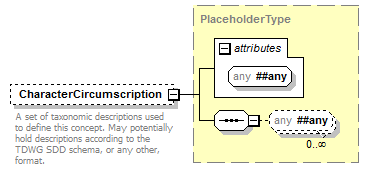 |
||||||||
| namespace | http://www.tdwg.org/schemas/tcs/1.01 | ||||||||
| type | PlaceholderType | ||||||||
| properties |
|
||||||||
| attributes |
|
||||||||
| annotation |
|
||||||||
| source | <xs:element name="CharacterCircumscription" type="PlaceholderType" minOccurs="0"> <xs:annotation> <xs:documentation>A set of taxonomic descriptions used to define this concept. May potentially hold descriptions according to the TDWG SDD schema, or any other, format.</xs:documentation> </xs:annotation> </xs:element> |
element DataSet/TaxonConcepts/TaxonConcept/ProviderLink
| diagram |  |
||||||||
| namespace | http://www.tdwg.org/schemas/tcs/1.01 | ||||||||
| type | xs:string | ||||||||
| properties |
|
||||||||
| source | <xs:element name="ProviderLink" type="xs:string" minOccurs="0"/> |
element DataSet/TaxonConcepts/TaxonConcept/ProviderSpecificData
| diagram |  |
||||||||
| namespace | http://www.tdwg.org/schemas/tcs/1.01 | ||||||||
| type | PlaceholderType | ||||||||
| properties |
|
||||||||
| attributes |
|
||||||||
| annotation |
|
||||||||
| source | <xs:element name="ProviderSpecificData" type="PlaceholderType" minOccurs="0"> <xs:annotation> <xs:documentation>Mechanism to allow for the extension of the schema for specific applications.</xs:documentation> </xs:annotation> </xs:element> |
element DataSet/TaxonRelationshipAssertions
| diagram |  |
||||||||
| namespace | http://www.tdwg.org/schemas/tcs/1.01 | ||||||||
| properties |
|
||||||||
| children | TaxonRelationshipAssertion | ||||||||
| annotation |
|
||||||||
| source | <xs:element name="TaxonRelationshipAssertions" minOccurs="0"> <xs:annotation> <xs:documentation>Container for taxonomic assertion records.</xs:documentation> </xs:annotation> <xs:complexType> <xs:sequence> <xs:element name="TaxonRelationshipAssertion" minOccurs="0" maxOccurs="unbounded"> <xs:annotation> <xs:documentation>Relationships between two concepts which are not part of the original definition of either of these concepts; possibly by a third party. </xs:documentation> </xs:annotation> <xs:complexType> <xs:complexContent> <xs:extension base="RelationshipType"> <xs:sequence minOccurs="0"> <xs:element name="AccordingTo" type="AccordingToType"> <xs:annotation> <xs:documentation>Information about the authorship of the asserted relationship.</xs:documentation> </xs:annotation> </xs:element> <xs:element name="FromTaxonConcept"> <xs:annotation> <xs:documentation>Starting point of the directed relationship. A reference to a TaxonConcept.</xs:documentation> </xs:annotation> <xs:complexType> <xs:complexContent> <xs:extension base="ReferenceType"/> </xs:complexContent> </xs:complexType> </xs:element> <xs:element name="ToTaxonConcept"> <xs:annotation> <xs:documentation>End point of the directed relationship. A reference to a TaxonConcept.</xs:documentation> </xs:annotation> <xs:complexType> <xs:complexContent> <xs:extension base="ReferenceType"/> </xs:complexContent> </xs:complexType> </xs:element> </xs:sequence> <xs:attribute name="id" type="xs:token" use="required"> <xs:annotation> <xs:documentation>Unique identifier (key) of the element. It can be local to the data set or a GUID in which case it can be resolved to a location on the internet where the record is stored. </xs:documentation> </xs:annotation> </xs:attribute> </xs:extension> </xs:complexContent> </xs:complexType> </xs:element> </xs:sequence> </xs:complexType> </xs:element> |
element DataSet/TaxonRelationshipAssertions/TaxonRelationshipAssertion
| diagram | 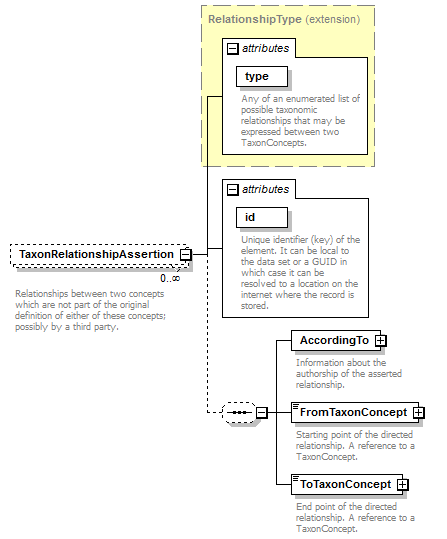 |
||||||||||||||||||||||
| namespace | http://www.tdwg.org/schemas/tcs/1.01 | ||||||||||||||||||||||
| type | extension of RelationshipType | ||||||||||||||||||||||
| properties |
|
||||||||||||||||||||||
| children | AccordingTo FromTaxonConcept ToTaxonConcept | ||||||||||||||||||||||
| attributes |
|
||||||||||||||||||||||
| annotation |
|
||||||||||||||||||||||
| source | <xs:element name="TaxonRelationshipAssertion" minOccurs="0" maxOccurs="unbounded"> <xs:annotation> <xs:documentation>Relationships between two concepts which are not part of the original definition of either of these concepts; possibly by a third party. </xs:documentation> </xs:annotation> <xs:complexType> <xs:complexContent> <xs:extension base="RelationshipType"> <xs:sequence minOccurs="0"> <xs:element name="AccordingTo" type="AccordingToType"> <xs:annotation> <xs:documentation>Information about the authorship of the asserted relationship.</xs:documentation> </xs:annotation> </xs:element> <xs:element name="FromTaxonConcept"> <xs:annotation> <xs:documentation>Starting point of the directed relationship. A reference to a TaxonConcept.</xs:documentation> </xs:annotation> <xs:complexType> <xs:complexContent> <xs:extension base="ReferenceType"/> </xs:complexContent> </xs:complexType> </xs:element> <xs:element name="ToTaxonConcept"> <xs:annotation> <xs:documentation>End point of the directed relationship. A reference to a TaxonConcept.</xs:documentation> </xs:annotation> <xs:complexType> <xs:complexContent> <xs:extension base="ReferenceType"/> </xs:complexContent> </xs:complexType> </xs:element> </xs:sequence> <xs:attribute name="id" type="xs:token" use="required"> <xs:annotation> <xs:documentation>Unique identifier (key) of the element. It can be local to the data set or a GUID in which case it can be resolved to a location on the internet where the record is stored. </xs:documentation> </xs:annotation> </xs:attribute> </xs:extension> </xs:complexContent> </xs:complexType> </xs:element> |
attribute DataSet/TaxonRelationshipAssertions/TaxonRelationshipAssertion/@id
| type | xs:token | ||||
| properties |
|
||||
| annotation |
|
||||
| source | <xs:attribute name="id" type="xs:token" use="required"> <xs:annotation> <xs:documentation>Unique identifier (key) of the element. It can be local to the data set or a GUID in which case it can be resolved to a location on the internet where the record is stored. </xs:documentation> </xs:annotation> </xs:attribute> |
element DataSet/TaxonRelationshipAssertions/TaxonRelationshipAssertion/AccordingTo
| diagram |  |
||||
| namespace | http://www.tdwg.org/schemas/tcs/1.01 | ||||
| type | AccordingToType | ||||
| properties |
|
||||
| children | Simple AccordingToDetailed | ||||
| annotation |
|
||||
| source | <xs:element name="AccordingTo" type="AccordingToType"> <xs:annotation> <xs:documentation>Information about the authorship of the asserted relationship.</xs:documentation> </xs:annotation> </xs:element> |
element DataSet/TaxonRelationshipAssertions/TaxonRelationshipAssertion/FromTaxonConcept
| diagram | 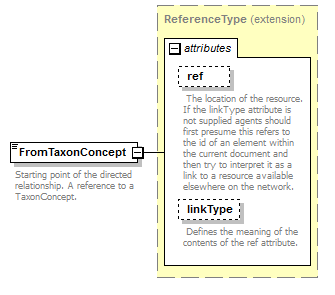 |
||||||||||||||||||||||
| namespace | http://www.tdwg.org/schemas/tcs/1.01 | ||||||||||||||||||||||
| type | extension of ReferenceType | ||||||||||||||||||||||
| properties |
|
||||||||||||||||||||||
| attributes |
|
||||||||||||||||||||||
| annotation |
|
||||||||||||||||||||||
| source | <xs:element name="FromTaxonConcept"> <xs:annotation> <xs:documentation>Starting point of the directed relationship. A reference to a TaxonConcept.</xs:documentation> </xs:annotation> <xs:complexType> <xs:complexContent> <xs:extension base="ReferenceType"/> </xs:complexContent> </xs:complexType> </xs:element> |
element DataSet/TaxonRelationshipAssertions/TaxonRelationshipAssertion/ToTaxonConcept
| diagram | 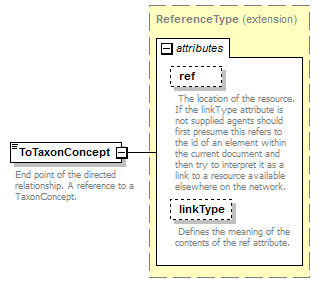 |
||||||||||||||||||||||
| namespace | http://www.tdwg.org/schemas/tcs/1.01 | ||||||||||||||||||||||
| type | extension of ReferenceType | ||||||||||||||||||||||
| properties |
|
||||||||||||||||||||||
| attributes |
|
||||||||||||||||||||||
| annotation |
|
||||||||||||||||||||||
| source | <xs:element name="ToTaxonConcept"> <xs:annotation> <xs:documentation>End point of the directed relationship. A reference to a TaxonConcept.</xs:documentation> </xs:annotation> <xs:complexType> <xs:complexContent> <xs:extension base="ReferenceType"/> </xs:complexContent> </xs:complexType> </xs:element> |
complexType AccordingToType
| diagram | 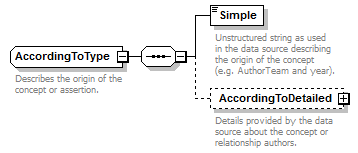 |
||
| namespace | http://www.tdwg.org/schemas/tcs/1.01 | ||
| children | Simple AccordingToDetailed | ||
| used by |
|
||
| annotation |
|
||
| source | <xs:complexType name="AccordingToType"> <xs:annotation> <xs:documentation>Describes the origin of the concept or assertion.</xs:documentation> </xs:annotation> <xs:sequence> <xs:element name="Simple" type="xs:string"> <xs:annotation> <xs:documentation>Unstructured string as used in the data source describing the origin of the concept (e.g. AuthorTeam and year).</xs:documentation> </xs:annotation> </xs:element> <xs:element name="AccordingToDetailed" minOccurs="0"> <xs:annotation> <xs:documentation>Details provided by the data source about the concept or relationship authors.</xs:documentation> </xs:annotation> <xs:complexType> <xs:sequence> <xs:element name="AuthorTeam" type="NameCitation" minOccurs="0"> <xs:annotation> <xs:documentation>This field should contain the authors of the concept. Note that the Simple sub element of this element is a signature field and should have the following content. For TCS signatures two situations are envisaged, one where concepts appear in printed publications and the other where concepts are published on-line. When representing a printed concept the field should contain the unabbreviated surnames of the authors in the order they appear in the publication separated by spaces. Initials and any punctuation marks should be omitted. If there are more than three authors only the first two author names should be included and they should be followed by the words “et al”. The full authorship of the concept will always be available via the PublishedIn element. Transliteration of names should be avoided unless they can't be represented in UTF-8 encoding. If the concept is being published on line, and does not exist in a paper form, then the DNS name of the institution publishing the concept should be used. A policy should be formulated for how many sub-domains should be cited and this should be stuck to. It is recommended that the www sub-domain should not be used (e.g. ipni.org not www.ipni.org). These DNS names are not expected to resolve to anything now or in future and so artificial sub-domains could be created to represent publishing authorities within larger organisation if required. If the concept is version sensitive then the DNS name should be followed by a space and then the versioning information. </xs:documentation> </xs:annotation> </xs:element> <xs:element name="PublishedIn" type="ReferenceType" minOccurs="0"> <xs:annotation> <xs:documentation>Reference ID or GUID of the original publication in which the concept or relationship was introduced. </xs:documentation> </xs:annotation> </xs:element> <xs:element name="MicroReference" type="xs:string" minOccurs="0"> <xs:annotation> <xs:documentation> This is an additional qualification to the reference given by 'PublishedIn'. It for holding things such as "page 34" or "tab. 67" etc. </xs:documentation> </xs:annotation> </xs:element> </xs:sequence> </xs:complexType> </xs:element> </xs:sequence> </xs:complexType> |
element AccordingToType/Simple
| diagram |  |
||||
| namespace | http://www.tdwg.org/schemas/tcs/1.01 | ||||
| type | xs:string | ||||
| properties |
|
||||
| annotation |
|
||||
| source | <xs:element name="Simple" type="xs:string"> <xs:annotation> <xs:documentation>Unstructured string as used in the data source describing the origin of the concept (e.g. AuthorTeam and year).</xs:documentation> </xs:annotation> </xs:element> |
element AccordingToType/AccordingToDetailed
| diagram |  |
||||||||
| namespace | http://www.tdwg.org/schemas/tcs/1.01 | ||||||||
| properties |
|
||||||||
| children | AuthorTeam PublishedIn MicroReference | ||||||||
| annotation |
|
||||||||
| source | <xs:element name="AccordingToDetailed" minOccurs="0"> <xs:annotation> <xs:documentation>Details provided by the data source about the concept or relationship authors.</xs:documentation> </xs:annotation> <xs:complexType> <xs:sequence> <xs:element name="AuthorTeam" type="NameCitation" minOccurs="0"> <xs:annotation> <xs:documentation>This field should contain the authors of the concept. Note that the Simple sub element of this element is a signature field and should have the following content. For TCS signatures two situations are envisaged, one where concepts appear in printed publications and the other where concepts are published on-line. When representing a printed concept the field should contain the unabbreviated surnames of the authors in the order they appear in the publication separated by spaces. Initials and any punctuation marks should be omitted. If there are more than three authors only the first two author names should be included and they should be followed by the words “et al”. The full authorship of the concept will always be available via the PublishedIn element. Transliteration of names should be avoided unless they can't be represented in UTF-8 encoding. If the concept is being published on line, and does not exist in a paper form, then the DNS name of the institution publishing the concept should be used. A policy should be formulated for how many sub-domains should be cited and this should be stuck to. It is recommended that the www sub-domain should not be used (e.g. ipni.org not www.ipni.org). These DNS names are not expected to resolve to anything now or in future and so artificial sub-domains could be created to represent publishing authorities within larger organisation if required. If the concept is version sensitive then the DNS name should be followed by a space and then the versioning information. </xs:documentation> </xs:annotation> </xs:element> <xs:element name="PublishedIn" type="ReferenceType" minOccurs="0"> <xs:annotation> <xs:documentation>Reference ID or GUID of the original publication in which the concept or relationship was introduced. </xs:documentation> </xs:annotation> </xs:element> <xs:element name="MicroReference" type="xs:string" minOccurs="0"> <xs:annotation> <xs:documentation> This is an additional qualification to the reference given by 'PublishedIn'. It for holding things such as "page 34" or "tab. 67" etc. </xs:documentation> </xs:annotation> </xs:element> </xs:sequence> </xs:complexType> </xs:element> |
element AccordingToType/AccordingToDetailed/AuthorTeam
| diagram |  |
||||||||
| namespace | http://www.tdwg.org/schemas/tcs/1.01 | ||||||||
| type | NameCitation | ||||||||
| properties |
|
||||||||
| children | Simple Year Authors | ||||||||
| annotation |
|
||||||||
| source | <xs:element name="AuthorTeam" type="NameCitation" minOccurs="0"> <xs:annotation> <xs:documentation>This field should contain the authors of the concept. Note that the Simple sub element of this element is a signature field and should have the following content. For TCS signatures two situations are envisaged, one where concepts appear in printed publications and the other where concepts are published on-line. When representing a printed concept the field should contain the unabbreviated surnames of the authors in the order they appear in the publication separated by spaces. Initials and any punctuation marks should be omitted. If there are more than three authors only the first two author names should be included and they should be followed by the words “et al”. The full authorship of the concept will always be available via the PublishedIn element. Transliteration of names should be avoided unless they can't be represented in UTF-8 encoding. If the concept is being published on line, and does not exist in a paper form, then the DNS name of the institution publishing the concept should be used. A policy should be formulated for how many sub-domains should be cited and this should be stuck to. It is recommended that the www sub-domain should not be used (e.g. ipni.org not www.ipni.org). These DNS names are not expected to resolve to anything now or in future and so artificial sub-domains could be created to represent publishing authorities within larger organisation if required. If the concept is version sensitive then the DNS name should be followed by a space and then the versioning information. </xs:documentation> </xs:annotation> </xs:element> |
element AccordingToType/AccordingToDetailed/PublishedIn
| diagram | 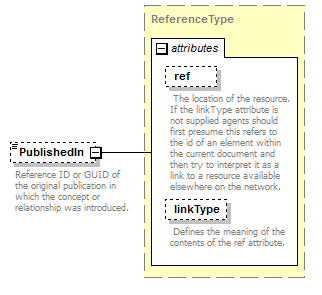 |
||||||||||||||||||||||
| namespace | http://www.tdwg.org/schemas/tcs/1.01 | ||||||||||||||||||||||
| type | ReferenceType | ||||||||||||||||||||||
| properties |
|
||||||||||||||||||||||
| attributes |
|
||||||||||||||||||||||
| annotation |
|
||||||||||||||||||||||
| source | <xs:element name="PublishedIn" type="ReferenceType" minOccurs="0"> <xs:annotation> <xs:documentation>Reference ID or GUID of the original publication in which the concept or relationship was introduced. </xs:documentation> </xs:annotation> </xs:element> |
element AccordingToType/AccordingToDetailed/MicroReference
| diagram |  |
||||||||
| namespace | http://www.tdwg.org/schemas/tcs/1.01 | ||||||||
| type | xs:string | ||||||||
| properties |
|
||||||||
| annotation |
|
||||||||
| source | <xs:element name="MicroReference" type="xs:string" minOccurs="0"> <xs:annotation> <xs:documentation> This is an additional qualification to the reference given by 'PublishedIn'. It for holding things such as "page 34" or "tab. 67" etc. </xs:documentation> </xs:annotation> </xs:element> |
complexType AgentNames
| diagram | 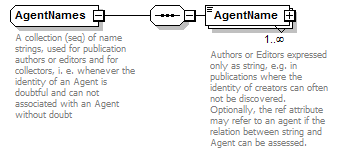 |
||
| namespace | http://www.tdwg.org/schemas/tcs/1.01 | ||
| children | AgentName | ||
| used by |
|
||
| annotation |
|
||
| source | <xs:complexType name="AgentNames"> <xs:annotation> <xs:documentation>A collection (seq) of name strings, used for publication authors or editors and for collectors, i. e. whenever the identity of an Agent is doubtful and can not associated with an Agent without doubt</xs:documentation> </xs:annotation> <xs:sequence> <xs:element name="AgentName" maxOccurs="unbounded"> <xs:annotation> <xs:documentation>Authors or Editors expressed only as string, e.g. in publications where the identity of creators can often not be discovered. Optionally, the ref attribute may refer to an agent if the relation between string and Agent can be assessed.</xs:documentation> </xs:annotation> <xs:complexType> <xs:complexContent> <xs:extension base="ReferenceType"> <xs:attribute name="role" use="optional"> <xs:simpleType> <xs:restriction base="xs:Name"> <xs:enumeration value="ex"/> <xs:enumeration value="sanctioning"/> </xs:restriction> </xs:simpleType> </xs:attribute> </xs:extension> </xs:complexContent> </xs:complexType> </xs:element> </xs:sequence> </xs:complexType> |
element AgentNames/AgentName
| diagram | 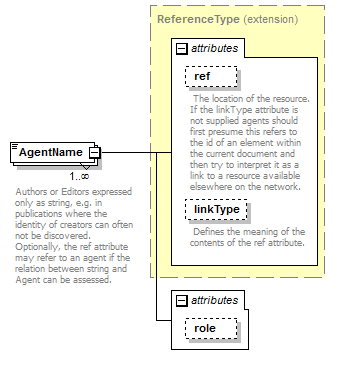 |
||||||||||||||||||||||||||||
| namespace | http://www.tdwg.org/schemas/tcs/1.01 | ||||||||||||||||||||||||||||
| type | extension of ReferenceType | ||||||||||||||||||||||||||||
| properties |
|
||||||||||||||||||||||||||||
| attributes |
|
||||||||||||||||||||||||||||
| annotation |
|
||||||||||||||||||||||||||||
| source | <xs:element name="AgentName" maxOccurs="unbounded"> <xs:annotation> <xs:documentation>Authors or Editors expressed only as string, e.g. in publications where the identity of creators can often not be discovered. Optionally, the ref attribute may refer to an agent if the relation between string and Agent can be assessed.</xs:documentation> </xs:annotation> <xs:complexType> <xs:complexContent> <xs:extension base="ReferenceType"> <xs:attribute name="role" use="optional"> <xs:simpleType> <xs:restriction base="xs:Name"> <xs:enumeration value="ex"/> <xs:enumeration value="sanctioning"/> </xs:restriction> </xs:simpleType> </xs:attribute> </xs:extension> </xs:complexContent> </xs:complexType> </xs:element> |
attribute AgentNames/AgentName/@role
| type | restriction of xs:Name | |||||||||
| properties |
|
|||||||||
| facets |
|
|||||||||
| source | <xs:attribute name="role" use="optional"> <xs:simpleType> <xs:restriction base="xs:Name"> <xs:enumeration value="ex"/> <xs:enumeration value="sanctioning"/> </xs:restriction> </xs:simpleType> </xs:attribute> |
complexType CanonicalAuthorship
| diagram | 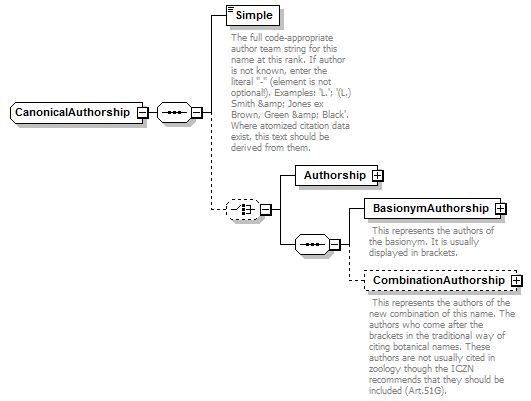 |
||
| namespace | http://www.tdwg.org/schemas/tcs/1.01 | ||
| children | Simple Authorship BasionymAuthorship CombinationAuthorship | ||
| used by |
|
||
| source | <xs:complexType name="CanonicalAuthorship"> <xs:sequence> <xs:element name="Simple" type="xs:string"> <xs:annotation> <xs:documentation>The full code-appropriate author team string for this name at this rank. If author is not known, enter the literal "-" (element is not optional!). Examples: 'L.'; '(L.) Smith & Jones ex Brown, Green & Black'. Where atomized citation data exist, this text should be derived from them.</xs:documentation> </xs:annotation> </xs:element> <xs:choice minOccurs="0"> <xs:element name="Authorship" type="NameCitation"/> <xs:sequence> <xs:element name="BasionymAuthorship" type="NameCitation"> <xs:annotation> <xs:documentation> This represents the authors of the basionym. It is usually displayed in brackets. </xs:documentation> </xs:annotation> </xs:element> <xs:element name="CombinationAuthorship" type="NameCitation" minOccurs="0"> <xs:annotation> <xs:documentation> This represents the authors of the new combination of this name. The authors who come after the brackets in the traditional way of citing botanical names. These authors are not usually cited in zoology though the ICZN recommends that they should be included (Art.51G). </xs:documentation> </xs:annotation> </xs:element> </xs:sequence> </xs:choice> </xs:sequence> </xs:complexType> |
element CanonicalAuthorship/Simple
| diagram |  |
||||
| namespace | http://www.tdwg.org/schemas/tcs/1.01 | ||||
| type | xs:string | ||||
| properties |
|
||||
| annotation |
|
||||
| source | <xs:element name="Simple" type="xs:string"> <xs:annotation> <xs:documentation>The full code-appropriate author team string for this name at this rank. If author is not known, enter the literal "-" (element is not optional!). Examples: 'L.'; '(L.) Smith & Jones ex Brown, Green & Black'. Where atomized citation data exist, this text should be derived from them.</xs:documentation> </xs:annotation> </xs:element> |
element CanonicalAuthorship/Authorship
| diagram | 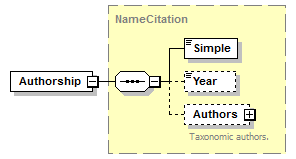 |
||||
| namespace | http://www.tdwg.org/schemas/tcs/1.01 | ||||
| type | NameCitation | ||||
| properties |
|
||||
| children | Simple Year Authors | ||||
| source | <xs:element name="Authorship" type="NameCitation"/> |
element CanonicalAuthorship/BasionymAuthorship
| diagram | 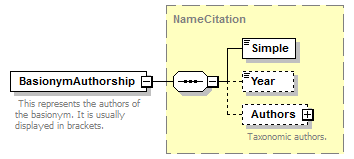 |
||||
| namespace | http://www.tdwg.org/schemas/tcs/1.01 | ||||
| type | NameCitation | ||||
| properties |
|
||||
| children | Simple Year Authors | ||||
| annotation |
|
||||
| source | <xs:element name="BasionymAuthorship" type="NameCitation"> <xs:annotation> <xs:documentation> This represents the authors of the basionym. It is usually displayed in brackets. </xs:documentation> </xs:annotation> </xs:element> |
element CanonicalAuthorship/CombinationAuthorship
| diagram | 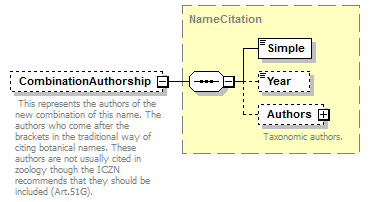 |
||||||||
| namespace | http://www.tdwg.org/schemas/tcs/1.01 | ||||||||
| type | NameCitation | ||||||||
| properties |
|
||||||||
| children | Simple Year Authors | ||||||||
| annotation |
|
||||||||
| source | <xs:element name="CombinationAuthorship" type="NameCitation" minOccurs="0"> <xs:annotation> <xs:documentation> This represents the authors of the new combination of this name. The authors who come after the brackets in the traditional way of citing botanical names. These authors are not usually cited in zoology though the ICZN recommends that they should be included (Art.51G). </xs:documentation> </xs:annotation> </xs:element> |
complexType CanonicalName
| diagram |  |
||
| namespace | http://www.tdwg.org/schemas/tcs/1.01 | ||
| children | Simple Uninomial Genus InfragenericEpithet SpecificEpithet InfraspecificEpithet CultivarNameGroup | ||
| used by |
|
||
| source | <xs:complexType name="CanonicalName"> <xs:sequence> <xs:element name="Simple" type="xs:string"> <xs:annotation> <xs:documentation> A label containing the canonical name without authors. For the purposes of a TCS signature this field should contain nothing but the text strings representing the name itself with no indication of rank or any other qualifiers. </xs:documentation> </xs:annotation> </xs:element> <xs:choice minOccurs="0"> <xs:element name="Uninomial" type="xs:string"> <xs:annotation> <xs:documentation>Family, genus, infrafamilial or suprafamilial name</xs:documentation> </xs:annotation> </xs:element> <xs:sequence> <xs:element name="Genus" type="ReferenceType"> <xs:annotation> <xs:documentation> The name of the genus. If this name is below the rank of genus then this may also be reference to a TaxonName that contains the name of the genus. </xs:documentation> </xs:annotation> </xs:element> <xs:choice> <xs:element name="InfragenericEpithet" type="xs:string"/> <xs:sequence> <xs:element name="SpecificEpithet" type="xs:string"> <xs:annotation> <xs:documentation> The specific epithet for the name. If this is subspecific taxon then this may be a link to another TaxonName that contains the species. </xs:documentation> </xs:annotation> </xs:element> <xs:element name="InfraspecificEpithet" type="xs:string" minOccurs="0"/> </xs:sequence> </xs:choice> </xs:sequence> </xs:choice> <xs:element name="CultivarNameGroup" type="xs:string" minOccurs="0"> <xs:annotation> <xs:documentation> The name of the Cultivar, Cultivar Group, grex, convar or graft chimera. Under the ICNCP. Just include here the string of the name. i.e. omit the single quotes around cultivar names, the word Group that denotes cultivar group and the + sign used in chimeras. These symbols can be added in later on the basis of the rank of the name. The user of Group for example is language dependant. </xs:documentation> </xs:annotation> </xs:element> </xs:sequence> </xs:complexType> |
element CanonicalName/Simple
| diagram |  |
||||
| namespace | http://www.tdwg.org/schemas/tcs/1.01 | ||||
| type | xs:string | ||||
| properties |
|
||||
| annotation |
|
||||
| source | <xs:element name="Simple" type="xs:string"> <xs:annotation> <xs:documentation> A label containing the canonical name without authors. For the purposes of a TCS signature this field should contain nothing but the text strings representing the name itself with no indication of rank or any other qualifiers. </xs:documentation> </xs:annotation> </xs:element> |
element CanonicalName/Uninomial
| diagram |  |
||||
| namespace | http://www.tdwg.org/schemas/tcs/1.01 | ||||
| type | xs:string | ||||
| properties |
|
||||
| annotation |
|
||||
| source | <xs:element name="Uninomial" type="xs:string"> <xs:annotation> <xs:documentation>Family, genus, infrafamilial or suprafamilial name</xs:documentation> </xs:annotation> </xs:element> |
element CanonicalName/Genus
| diagram |  |
||||||||||||||||||||||
| namespace | http://www.tdwg.org/schemas/tcs/1.01 | ||||||||||||||||||||||
| type | ReferenceType | ||||||||||||||||||||||
| properties |
|
||||||||||||||||||||||
| attributes |
|
||||||||||||||||||||||
| annotation |
|
||||||||||||||||||||||
| source | <xs:element name="Genus" type="ReferenceType"> <xs:annotation> <xs:documentation> The name of the genus. If this name is below the rank of genus then this may also be reference to a TaxonName that contains the name of the genus. </xs:documentation> </xs:annotation> </xs:element> |
element CanonicalName/InfragenericEpithet
| diagram |  |
||||
| namespace | http://www.tdwg.org/schemas/tcs/1.01 | ||||
| type | xs:string | ||||
| properties |
|
||||
| source | <xs:element name="InfragenericEpithet" type="xs:string"/> |
element CanonicalName/SpecificEpithet
| diagram |  |
||||
| namespace | http://www.tdwg.org/schemas/tcs/1.01 | ||||
| type | xs:string | ||||
| properties |
|
||||
| annotation |
|
||||
| source | <xs:element name="SpecificEpithet" type="xs:string"> <xs:annotation> <xs:documentation> The specific epithet for the name. If this is subspecific taxon then this may be a link to another TaxonName that contains the species. </xs:documentation> </xs:annotation> </xs:element> |
element CanonicalName/InfraspecificEpithet
| diagram |  |
||||||||
| namespace | http://www.tdwg.org/schemas/tcs/1.01 | ||||||||
| type | xs:string | ||||||||
| properties |
|
||||||||
| source | <xs:element name="InfraspecificEpithet" type="xs:string" minOccurs="0"/> |
element CanonicalName/CultivarNameGroup
| diagram |  |
||||||||
| namespace | http://www.tdwg.org/schemas/tcs/1.01 | ||||||||
| type | xs:string | ||||||||
| properties |
|
||||||||
| annotation |
|
||||||||
| source | <xs:element name="CultivarNameGroup" type="xs:string" minOccurs="0"> <xs:annotation> <xs:documentation> The name of the Cultivar, Cultivar Group, grex, convar or graft chimera. Under the ICNCP. Just include here the string of the name. i.e. omit the single quotes around cultivar names, the word Group that denotes cultivar group and the + sign used in chimeras. These symbols can be added in later on the basis of the rank of the name. The user of Group for example is language dependant. </xs:documentation> </xs:annotation> </xs:element> |
complexType NameCitation
| diagram | 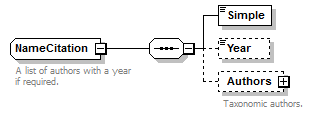 |
||
| namespace | http://www.tdwg.org/schemas/tcs/1.01 | ||
| children | Simple Year Authors | ||
| used by |
|
||
| annotation |
|
||
| source | <xs:complexType name="NameCitation"> <xs:annotation> <xs:documentation>A list of authors with a year if required.</xs:documentation> </xs:annotation> <xs:sequence> <xs:element name="Simple" type="xs:string"/> <xs:element name="Year" type="xs:gYear" minOccurs="0"/> <xs:element name="Authors" type="AgentNames" minOccurs="0"> <xs:annotation> <xs:documentation>Taxonomic authors. </xs:documentation> </xs:annotation> </xs:element> </xs:sequence> </xs:complexType> |
element NameCitation/Simple
| diagram |  |
||||
| namespace | http://www.tdwg.org/schemas/tcs/1.01 | ||||
| type | xs:string | ||||
| properties |
|
||||
| source | <xs:element name="Simple" type="xs:string"/> |
element NameCitation/Year
| diagram |  |
||||||||
| namespace | http://www.tdwg.org/schemas/tcs/1.01 | ||||||||
| type | xs:gYear | ||||||||
| properties |
|
||||||||
| source | <xs:element name="Year" type="xs:gYear" minOccurs="0"/> |
element NameCitation/Authors
| diagram | 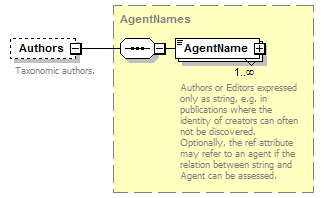 |
||||||||
| namespace | http://www.tdwg.org/schemas/tcs/1.01 | ||||||||
| type | AgentNames | ||||||||
| properties |
|
||||||||
| children | AgentName | ||||||||
| annotation |
|
||||||||
| source | <xs:element name="Authors" type="AgentNames" minOccurs="0"> <xs:annotation> <xs:documentation>Taxonomic authors. </xs:documentation> </xs:annotation> </xs:element> |
complexType NomenclaturalNoteType
| diagram | 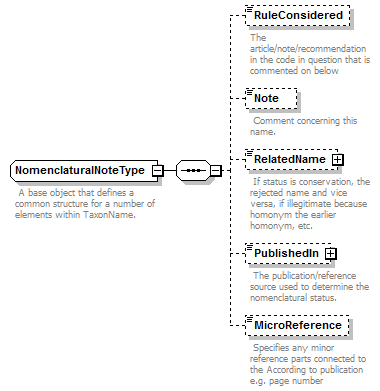 |
||
| namespace | http://www.tdwg.org/schemas/tcs/1.01 | ||
| children | RuleConsidered Note RelatedName PublishedIn MicroReference | ||
| used by | |||
| annotation |
|
||
| source | <xs:complexType name="NomenclaturalNoteType"> <xs:annotation> <xs:documentation> A base object that defines a common structure for a number of elements within TaxonName. </xs:documentation> </xs:annotation> <xs:sequence> <xs:element name="RuleConsidered" type="xs:string" minOccurs="0"> <xs:annotation> <xs:documentation>The article/note/recommendation in the code in question that is commented on below</xs:documentation> </xs:annotation> </xs:element> <xs:element name="Note" type="xs:string" minOccurs="0"> <xs:annotation> <xs:documentation> Comment concerning this name. </xs:documentation> </xs:annotation> </xs:element> <xs:element name="RelatedName" type="ReferenceType" minOccurs="0"> <xs:annotation> <xs:documentation> If status is conservation, the rejected name and vice versa, if illegitimate because homonym the earlier homonym, etc. </xs:documentation> </xs:annotation> </xs:element> <xs:element name="PublishedIn" type="ReferenceType" minOccurs="0"> <xs:annotation> <xs:documentation> The publication/reference source used to determine the nomenclatural status. </xs:documentation> </xs:annotation> </xs:element> <xs:element name="MicroReference" type="xs:string" minOccurs="0"> <xs:annotation> <xs:documentation> Specifies any minor reference parts connected to the According to publication e.g. page number </xs:documentation> </xs:annotation> </xs:element> </xs:sequence> </xs:complexType> |
element NomenclaturalNoteType/RuleConsidered
| diagram |  |
||||||||
| namespace | http://www.tdwg.org/schemas/tcs/1.01 | ||||||||
| type | xs:string | ||||||||
| properties |
|
||||||||
| annotation |
|
||||||||
| source | <xs:element name="RuleConsidered" type="xs:string" minOccurs="0"> <xs:annotation> <xs:documentation>The article/note/recommendation in the code in question that is commented on below</xs:documentation> </xs:annotation> </xs:element> |
element NomenclaturalNoteType/Note
| diagram |  |
||||||||
| namespace | http://www.tdwg.org/schemas/tcs/1.01 | ||||||||
| type | xs:string | ||||||||
| properties |
|
||||||||
| annotation |
|
||||||||
| source | <xs:element name="Note" type="xs:string" minOccurs="0"> <xs:annotation> <xs:documentation> Comment concerning this name. </xs:documentation> </xs:annotation> </xs:element> |
element NomenclaturalNoteType/RelatedName
| diagram | 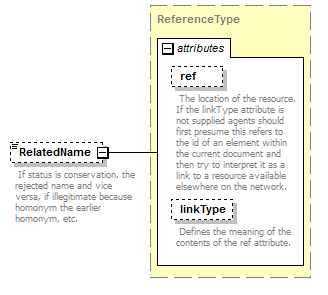 |
||||||||||||||||||||||
| namespace | http://www.tdwg.org/schemas/tcs/1.01 | ||||||||||||||||||||||
| type | ReferenceType | ||||||||||||||||||||||
| properties |
|
||||||||||||||||||||||
| attributes |
|
||||||||||||||||||||||
| annotation |
|
||||||||||||||||||||||
| source | <xs:element name="RelatedName" type="ReferenceType" minOccurs="0"> <xs:annotation> <xs:documentation> If status is conservation, the rejected name and vice versa, if illegitimate because homonym the earlier homonym, etc. </xs:documentation> </xs:annotation> </xs:element> |
element NomenclaturalNoteType/PublishedIn
| diagram | 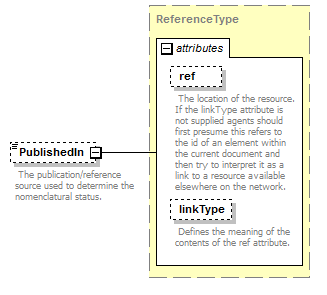 |
||||||||||||||||||||||
| namespace | http://www.tdwg.org/schemas/tcs/1.01 | ||||||||||||||||||||||
| type | ReferenceType | ||||||||||||||||||||||
| properties |
|
||||||||||||||||||||||
| attributes |
|
||||||||||||||||||||||
| annotation |
|
||||||||||||||||||||||
| source | <xs:element name="PublishedIn" type="ReferenceType" minOccurs="0"> <xs:annotation> <xs:documentation> The publication/reference source used to determine the nomenclatural status. </xs:documentation> </xs:annotation> </xs:element> |
element NomenclaturalNoteType/MicroReference
| diagram | 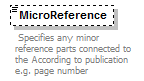 |
||||||||
| namespace | http://www.tdwg.org/schemas/tcs/1.01 | ||||||||
| type | xs:string | ||||||||
| properties |
|
||||||||
| annotation |
|
||||||||
| source | <xs:element name="MicroReference" type="xs:string" minOccurs="0"> <xs:annotation> <xs:documentation> Specifies any minor reference parts connected to the According to publication e.g. page number </xs:documentation> </xs:annotation> </xs:element> |
complexType PlaceholderType
| diagram |  |
||||||
| namespace | http://www.tdwg.org/schemas/tcs/1.01 | ||||||
| used by | |||||||
| attributes |
|
||||||
| annotation |
|
||||||
| source | <xs:complexType name="PlaceholderType"> <xs:annotation> <xs:documentation>Placeholder for other schema standards</xs:documentation> </xs:annotation> <xs:sequence> <xs:any namespace="##any" processContents="lax" minOccurs="0" maxOccurs="unbounded"/> </xs:sequence> <xs:anyAttribute namespace="##any" processContents="lax"/> </xs:complexType> |
complexType ReferenceType
| diagram | 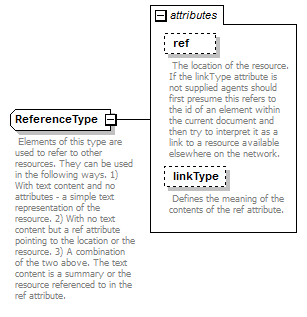 |
||||||||||||||||||||||
| namespace | http://www.tdwg.org/schemas/tcs/1.01 | ||||||||||||||||||||||
| properties |
|
||||||||||||||||||||||
| used by | |||||||||||||||||||||||
| attributes |
|
||||||||||||||||||||||
| annotation |
|
||||||||||||||||||||||
| source | <xs:complexType name="ReferenceType" mixed="true"> <xs:annotation> <xs:documentation> Elements of this type are used to refer to other resources. They can be used in the following ways. 1) With text content and no attributes - a simple text representation of the resource. 2) With no text content but a ref attribute pointing to the location or the resource. 3) A combination of the two above. The text content is a summary or the resource referenced to in the ref attribute. </xs:documentation> </xs:annotation> <xs:attribute name="ref" type="xs:token" use="optional"> <xs:annotation> <xs:documentation> The location of the resource. If the linkType attribute is not supplied agents should first presume this refers to the id of an element within the current document and then try to interpret it as a link to a resource available elsewhere on the network. </xs:documentation> </xs:annotation> </xs:attribute> <xs:attribute name="linkType" use="optional" default="local"> <xs:annotation> <xs:documentation> Defines the meaning of the contents of the ref attribute. </xs:documentation> </xs:annotation> <xs:simpleType> <xs:restriction base="xs:string"> <xs:enumeration value="local"> <xs:annotation> <xs:documentation> This is the id of another element within this document. </xs:documentation> </xs:annotation> </xs:enumeration> <xs:enumeration value="external"> <xs:annotation> <xs:documentation> A link to a resource in another document. It is presumed that the target will be returned as a valid TCS instance. </xs:documentation> </xs:annotation> </xs:enumeration> <xs:enumeration value="other"> <xs:annotation> <xs:documentation> A link to another resource external to the current document that is not in TCS format. e.g. a jpeg, html page or pdf. </xs:documentation> </xs:annotation> </xs:enumeration> </xs:restriction> </xs:simpleType> </xs:attribute> </xs:complexType> |
attribute ReferenceType/@ref
| type | xs:token | ||||
| properties |
|
||||
| annotation |
|
||||
| source | <xs:attribute name="ref" type="xs:token" use="optional"> <xs:annotation> <xs:documentation> The location of the resource. If the linkType attribute is not supplied agents should first presume this refers to the id of an element within the current document and then try to interpret it as a link to a resource available elsewhere on the network. </xs:documentation> </xs:annotation> </xs:attribute> |
attribute ReferenceType/@linkType
| type | restriction of xs:string | ||||||||||||||||||
| properties |
|
||||||||||||||||||
| facets |
|
||||||||||||||||||
| annotation |
|
||||||||||||||||||
| source | <xs:attribute name="linkType" use="optional" default="local"> <xs:annotation> <xs:documentation> Defines the meaning of the contents of the ref attribute. </xs:documentation> </xs:annotation> <xs:simpleType> <xs:restriction base="xs:string"> <xs:enumeration value="local"> <xs:annotation> <xs:documentation> This is the id of another element within this document. </xs:documentation> </xs:annotation> </xs:enumeration> <xs:enumeration value="external"> <xs:annotation> <xs:documentation> A link to a resource in another document. It is presumed that the target will be returned as a valid TCS instance. </xs:documentation> </xs:annotation> </xs:enumeration> <xs:enumeration value="other"> <xs:annotation> <xs:documentation> A link to another resource external to the current document that is not in TCS format. e.g. a jpeg, html page or pdf. </xs:documentation> </xs:annotation> </xs:enumeration> </xs:restriction> </xs:simpleType> </xs:attribute> |
complexType RelationshipType
| diagram | 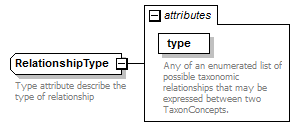 |
||||||||||||||
| namespace | http://www.tdwg.org/schemas/tcs/1.01 | ||||||||||||||
| used by |
|
||||||||||||||
| attributes |
|
||||||||||||||
| annotation |
|
||||||||||||||
| source | <xs:complexType name="RelationshipType"> <xs:annotation> <xs:documentation>Type attribute describe the type of relationship</xs:documentation> </xs:annotation> <xs:attribute name="type" use="required"> <xs:annotation> <xs:documentation>Any of an enumerated list of possible taxonomic relationships that may be expressed between two TaxonConcepts.</xs:documentation> </xs:annotation> <xs:simpleType> <xs:restriction base="xs:string"> <xs:enumeration value="is congruent to"> <xs:annotation> <xs:documentation> Set Relationship: The extent of Concept 1 is (essentially) identical to Concept 2 </xs:documentation> </xs:annotation> </xs:enumeration> <xs:enumeration value="is not congruent to"> <xs:annotation> <xs:documentation> Set Relationship: The extent of Concept 1 is not identical to Concept 2 </xs:documentation> </xs:annotation> </xs:enumeration> <xs:enumeration value="includes"> <xs:annotation> <xs:documentation> Set Relationship: Concept 2 is a subset of Concept 1 </xs:documentation> </xs:annotation> </xs:enumeration> <xs:enumeration value="does not include"> <xs:annotation> <xs:documentation> Set Relationship: Concept 2 is not a subset of Concept 1 </xs:documentation> </xs:annotation> </xs:enumeration> <xs:enumeration value="excludes"> <xs:annotation> <xs:documentation> Set Relationship: Concept 1 does not overlap or include Concept 2 </xs:documentation> </xs:annotation> </xs:enumeration> <xs:enumeration value="is included in"> <xs:annotation> <xs:documentation> Set Relationship: Concept 1 is a subset of Concept 2 </xs:documentation> </xs:annotation> </xs:enumeration> <xs:enumeration value="is not included in"> <xs:annotation> <xs:documentation> Set Relationship: Concept 1 is not a subset of Concept 2 </xs:documentation> </xs:annotation> </xs:enumeration> <xs:enumeration value="overlaps"> <xs:annotation> <xs:documentation> Set Relationship: Concepts 1 and 2 share members/children in common </xs:documentation> </xs:annotation> </xs:enumeration> <xs:enumeration value="does not overlap"> <xs:annotation> <xs:documentation> Set Relationship: Concepts 1 and 2 have no members/children in common </xs:documentation> </xs:annotation> </xs:enumeration> <xs:enumeration value="is child taxon of"> <xs:annotation> <xs:documentation> Hierarchical Relationship: Concept 1 is a member of lower taxonomic rank of Concept 2 </xs:documentation> </xs:annotation> </xs:enumeration> <xs:enumeration value="is parent taxon of"> <xs:annotation> <xs:documentation> Hierarchical Relationship: Taxon Concept 1 includes Concept 2 as a lower-ranked member. </xs:documentation> </xs:annotation> </xs:enumeration> <xs:enumeration value="is anamorph of"> <xs:annotation> <xs:documentation> Concept 1 is the asexual or mitotic reproductive stage in a pleomorphic life cycle in which Concept 2 is the teleomorph or meiotic reproductive stage . </xs:documentation> </xs:annotation> </xs:enumeration> <xs:enumeration value="is teleomorph of"> <xs:annotation> <xs:documentation> Concept 1 is the teleomorph or meiotic reproductive stage in a pleomorphic life cycle in which Concept 2 is the asexual or mitotic reproductive stage. </xs:documentation> </xs:annotation> </xs:enumeration> <xs:enumeration value="is second parent of"> <xs:annotation> <xs:documentation> Hybrid Relationship: Concept 1 is genetic parent (2) of Concept 2 . </xs:documentation> </xs:annotation> </xs:enumeration> <xs:enumeration value="is female parent of"> <xs:annotation> <xs:documentation> Hybrid Relationship: Concept 1 is genetic mother of Concept 2 </xs:documentation> </xs:annotation> </xs:enumeration> <xs:enumeration value="is first parent of"> <xs:annotation> <xs:documentation> Hybrid Relationship: Concept 1 is genetic parent (1) of Concept 2 </xs:documentation> </xs:annotation> </xs:enumeration> <xs:enumeration value="is male parent of"> <xs:annotation> <xs:documentation> Hybrid Relationship: Concept 1 is genetic father of Concept 2 </xs:documentation> </xs:annotation> </xs:enumeration> <xs:enumeration value="is hybrid parent of"> <xs:annotation> <xs:documentation> Hybrid Relationship: Concept 1 is genetic parent of Concept 2 </xs:documentation> </xs:annotation> </xs:enumeration> <xs:enumeration value="is hybrid child of"> <xs:annotation> <xs:documentation> Hybrid Relationship: Concept 2 is a genetic parent of Concept 1 </xs:documentation> </xs:annotation> </xs:enumeration> <xs:enumeration value="is ambiregnal of"> <xs:annotation> <xs:documentation> Concepts have only a single scientific name. This name is governed by a single nomenclatural code. If an organism falls into two codes (not a desirable or frequent situation) then two TaxonConcepts should be created and linked with this relationship. e.g. a concept for the organism as an animal and a concept of it as a plant. It is unlikely this will be widely used as a taxonomist would typically have a single view of which kingdom a organism is in. Also note that ambiregnal is used to imply different nomenclatural codes but there is not a precise relationship between codes and kingdoms.</xs:documentation> </xs:annotation> </xs:enumeration> <xs:enumeration value="is vernacular for"> <xs:annotation> <xs:documentation> The current concept is used as a vernacular concept, at least in part, for the target concept. This kind of relationship should not be used to express any form of set relationship (e.g. overlaps, is congruent with, includes). Consider using vernacular type relationships along with set type relationships to avoid any ambiguity. </xs:documentation> </xs:annotation> </xs:enumeration> <xs:enumeration value="has vernacular"> <xs:annotation> <xs:documentation> The target concept is used as a vernacular concept, at least in part, for the current concept. This kind of relationship should not be used to express any form of set relationship (e.g. overlaps, is congruent with, includes). Consider using vernacular type relationships along with set type relationships to avoid any ambiguity. </xs:documentation> </xs:annotation> </xs:enumeration> <xs:enumeration value="has synonym"> <xs:annotation> <xs:documentation> The target concept is considered a synonym of the current concept. This is an ambiguous relationship. It can mean: 1) a nomenclatural relationship where all that is implied is that the type of the target concept is included in the current circumscription. This is more precisely expressed as a SpecimenCircumscription (for heterotypic synonyms) or as TaxonName basionym relationships (for homotypic synonyms) 2) a concept relationship where some part of (or all of) the target concept is included in the current circumscription. This is more precisely expressed using the set relationships such as 'is congruent to'. This is intended for handling legacy data. </xs:documentation> </xs:annotation> </xs:enumeration> </xs:restriction> </xs:simpleType> </xs:attribute> </xs:complexType> |
attribute RelationshipType/@type
| type | restriction of xs:string | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| properties |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| facets |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| annotation |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| source | <xs:attribute name="type" use="required"> <xs:annotation> <xs:documentation>Any of an enumerated list of possible taxonomic relationships that may be expressed between two TaxonConcepts.</xs:documentation> </xs:annotation> <xs:simpleType> <xs:restriction base="xs:string"> <xs:enumeration value="is congruent to"> <xs:annotation> <xs:documentation> Set Relationship: The extent of Concept 1 is (essentially) identical to Concept 2 </xs:documentation> </xs:annotation> </xs:enumeration> <xs:enumeration value="is not congruent to"> <xs:annotation> <xs:documentation> Set Relationship: The extent of Concept 1 is not identical to Concept 2 </xs:documentation> </xs:annotation> </xs:enumeration> <xs:enumeration value="includes"> <xs:annotation> <xs:documentation> Set Relationship: Concept 2 is a subset of Concept 1 </xs:documentation> </xs:annotation> </xs:enumeration> <xs:enumeration value="does not include"> <xs:annotation> <xs:documentation> Set Relationship: Concept 2 is not a subset of Concept 1 </xs:documentation> </xs:annotation> </xs:enumeration> <xs:enumeration value="excludes"> <xs:annotation> <xs:documentation> Set Relationship: Concept 1 does not overlap or include Concept 2 </xs:documentation> </xs:annotation> </xs:enumeration> <xs:enumeration value="is included in"> <xs:annotation> <xs:documentation> Set Relationship: Concept 1 is a subset of Concept 2 </xs:documentation> </xs:annotation> </xs:enumeration> <xs:enumeration value="is not included in"> <xs:annotation> <xs:documentation> Set Relationship: Concept 1 is not a subset of Concept 2 </xs:documentation> </xs:annotation> </xs:enumeration> <xs:enumeration value="overlaps"> <xs:annotation> <xs:documentation> Set Relationship: Concepts 1 and 2 share members/children in common </xs:documentation> </xs:annotation> </xs:enumeration> <xs:enumeration value="does not overlap"> <xs:annotation> <xs:documentation> Set Relationship: Concepts 1 and 2 have no members/children in common </xs:documentation> </xs:annotation> </xs:enumeration> <xs:enumeration value="is child taxon of"> <xs:annotation> <xs:documentation> Hierarchical Relationship: Concept 1 is a member of lower taxonomic rank of Concept 2 </xs:documentation> </xs:annotation> </xs:enumeration> <xs:enumeration value="is parent taxon of"> <xs:annotation> <xs:documentation> Hierarchical Relationship: Taxon Concept 1 includes Concept 2 as a lower-ranked member. </xs:documentation> </xs:annotation> </xs:enumeration> <xs:enumeration value="is anamorph of"> <xs:annotation> <xs:documentation> Concept 1 is the asexual or mitotic reproductive stage in a pleomorphic life cycle in which Concept 2 is the teleomorph or meiotic reproductive stage . </xs:documentation> </xs:annotation> </xs:enumeration> <xs:enumeration value="is teleomorph of"> <xs:annotation> <xs:documentation> Concept 1 is the teleomorph or meiotic reproductive stage in a pleomorphic life cycle in which Concept 2 is the asexual or mitotic reproductive stage. </xs:documentation> </xs:annotation> </xs:enumeration> <xs:enumeration value="is second parent of"> <xs:annotation> <xs:documentation> Hybrid Relationship: Concept 1 is genetic parent (2) of Concept 2 . </xs:documentation> </xs:annotation> </xs:enumeration> <xs:enumeration value="is female parent of"> <xs:annotation> <xs:documentation> Hybrid Relationship: Concept 1 is genetic mother of Concept 2 </xs:documentation> </xs:annotation> </xs:enumeration> <xs:enumeration value="is first parent of"> <xs:annotation> <xs:documentation> Hybrid Relationship: Concept 1 is genetic parent (1) of Concept 2 </xs:documentation> </xs:annotation> </xs:enumeration> <xs:enumeration value="is male parent of"> <xs:annotation> <xs:documentation> Hybrid Relationship: Concept 1 is genetic father of Concept 2 </xs:documentation> </xs:annotation> </xs:enumeration> <xs:enumeration value="is hybrid parent of"> <xs:annotation> <xs:documentation> Hybrid Relationship: Concept 1 is genetic parent of Concept 2 </xs:documentation> </xs:annotation> </xs:enumeration> <xs:enumeration value="is hybrid child of"> <xs:annotation> <xs:documentation> Hybrid Relationship: Concept 2 is a genetic parent of Concept 1 </xs:documentation> </xs:annotation> </xs:enumeration> <xs:enumeration value="is ambiregnal of"> <xs:annotation> <xs:documentation> Concepts have only a single scientific name. This name is governed by a single nomenclatural code. If an organism falls into two codes (not a desirable or frequent situation) then two TaxonConcepts should be created and linked with this relationship. e.g. a concept for the organism as an animal and a concept of it as a plant. It is unlikely this will be widely used as a taxonomist would typically have a single view of which kingdom a organism is in. Also note that ambiregnal is used to imply different nomenclatural codes but there is not a precise relationship between codes and kingdoms.</xs:documentation> </xs:annotation> </xs:enumeration> <xs:enumeration value="is vernacular for"> <xs:annotation> <xs:documentation> The current concept is used as a vernacular concept, at least in part, for the target concept. This kind of relationship should not be used to express any form of set relationship (e.g. overlaps, is congruent with, includes). Consider using vernacular type relationships along with set type relationships to avoid any ambiguity. </xs:documentation> </xs:annotation> </xs:enumeration> <xs:enumeration value="has vernacular"> <xs:annotation> <xs:documentation> The target concept is used as a vernacular concept, at least in part, for the current concept. This kind of relationship should not be used to express any form of set relationship (e.g. overlaps, is congruent with, includes). Consider using vernacular type relationships along with set type relationships to avoid any ambiguity. </xs:documentation> </xs:annotation> </xs:enumeration> <xs:enumeration value="has synonym"> <xs:annotation> <xs:documentation> The target concept is considered a synonym of the current concept. This is an ambiguous relationship. It can mean: 1) a nomenclatural relationship where all that is implied is that the type of the target concept is included in the current circumscription. This is more precisely expressed as a SpecimenCircumscription (for heterotypic synonyms) or as TaxonName basionym relationships (for homotypic synonyms) 2) a concept relationship where some part of (or all of) the target concept is included in the current circumscription. This is more precisely expressed using the set relationships such as 'is congruent to'. This is intended for handling legacy data. </xs:documentation> </xs:annotation> </xs:enumeration> </xs:restriction> </xs:simpleType> </xs:attribute> |
complexType ScientificName
| diagram | 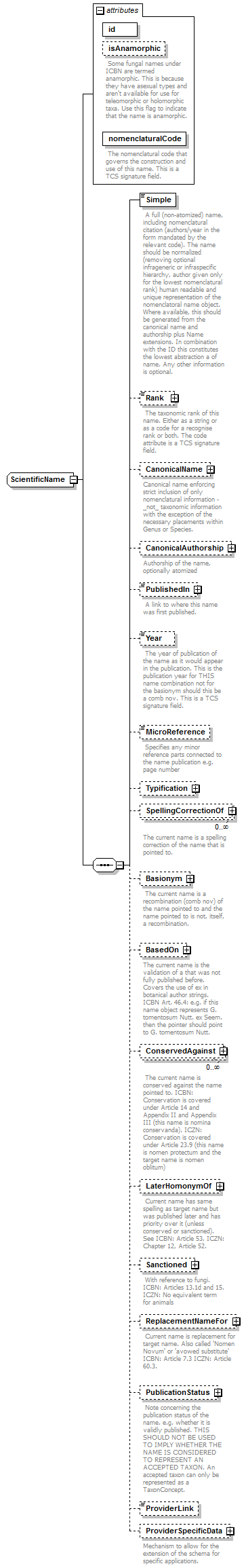 |
||||||||||||||||||||||||||||
| namespace | http://www.tdwg.org/schemas/tcs/1.01 | ||||||||||||||||||||||||||||
| children | Simple Rank CanonicalName CanonicalAuthorship PublishedIn Year MicroReference Typification SpellingCorrectionOf Basionym BasedOn ConservedAgainst LaterHomonymOf Sanctioned ReplacementNameFor PublicationStatus ProviderLink ProviderSpecificData | ||||||||||||||||||||||||||||
| used by |
|
||||||||||||||||||||||||||||
| attributes |
|
||||||||||||||||||||||||||||
| source | <xs:complexType name="ScientificName"> <xs:sequence> <xs:element name="Simple" type="xs:string"> <xs:annotation> <xs:documentation> A full (non-atomized) name, including nomenclatural citation (authors/year in the form mandated by the relevant code). The name should be normalized (removing optional infrageneric or infraspecific hierarchy, author given only for the lowest nomenclatural rank) human readable and unique representation of the nomenclatoral name object. Where available, this should be generated from the canonical name and authorship plus Name extensions. In combination with the ID this constitutes the lowest abstraction a of name. Any other information is optional. </xs:documentation> </xs:annotation> </xs:element> <xs:element name="Rank" type="TaxonomicRank" minOccurs="0"> <xs:annotation> <xs:documentation> The taxonomic rank of this name. Either as a string or as a code for a recognise rank or both. The code attribute is a TCS signature field. </xs:documentation> </xs:annotation> </xs:element> <xs:element name="CanonicalName" type="CanonicalName" minOccurs="0"> <xs:annotation> <xs:documentation>Canonical name enforcing strict inclusion of only nomenclatural information - _not_ taxonomic information with the exception of the necessary placements within Genus or Species. </xs:documentation> </xs:annotation> </xs:element> <xs:element name="CanonicalAuthorship" type="CanonicalAuthorship" minOccurs="0"> <xs:annotation> <xs:documentation>Authorship of the name, optionally atomized</xs:documentation> </xs:annotation> </xs:element> <xs:element name="PublishedIn" type="ReferenceType" minOccurs="0"> <xs:annotation> <xs:documentation> A link to where this name was first published. </xs:documentation> </xs:annotation> </xs:element> <xs:element name="Year" type="xs:gYear" minOccurs="0"> <xs:annotation> <xs:documentation> The year of publication of the name as it would appear in the publication. This is the publication year for THIS name combination not for the basionym should this be a comb nov. This is a TCS signature field. </xs:documentation> </xs:annotation> </xs:element> <xs:element name="MicroReference" type="xs:string" minOccurs="0"> <xs:annotation> <xs:documentation> Specifies any minor reference parts connected to the name publication e.g. page number </xs:documentation> </xs:annotation> </xs:element> <xs:element name="Typification" minOccurs="0"> <xs:complexType> <xs:sequence> <xs:element name="Simple" type="xs:string"/> <xs:choice> <xs:element name="TypeVouchers"> <xs:annotation> <xs:documentation> A container for type specimens and other vouchers. </xs:documentation> </xs:annotation> <xs:complexType> <xs:sequence> <xs:element name="TypeVoucher" maxOccurs="unbounded"> <xs:annotation> <xs:documentation>Names at species and below are typified by specimens.</xs:documentation> </xs:annotation> <xs:complexType> <xs:sequence> <xs:element name="VoucherReference" type="ReferenceType" minOccurs="0"> <xs:annotation> <xs:documentation> A reference to a type specimen for this name. </xs:documentation> </xs:annotation> </xs:element> <xs:element name="LectotypePublication" type="ReferenceType" minOccurs="0"> <xs:annotation> <xs:documentation> A reference to the publication where the lectotype was nominated. </xs:documentation> </xs:annotation> </xs:element> <xs:element name="LectotypeMicroReference" type="xs:string" minOccurs="0"> <xs:annotation> <xs:documentation> Specifies any minor reference parts connected to the lectotypification publication e.g. page number </xs:documentation> </xs:annotation> </xs:element> </xs:sequence> <xs:attribute name="typeOfType" type="NomenclaturalTypeStatusOfUnitsEnum"> <xs:annotation> <xs:documentation> The kind of type this specimen is e.g. paratype, isotype, holotype etc. </xs:documentation> </xs:annotation> </xs:attribute> </xs:complexType> </xs:element> </xs:sequence> </xs:complexType> </xs:element> <xs:element name="TypeName"> <xs:annotation> <xs:documentation>Names above species level are typified by the NAME of a lower taxon.</xs:documentation> </xs:annotation> <xs:complexType> <xs:sequence> <xs:element name="NameReference" type="ReferenceType" minOccurs="0"> <xs:annotation> <xs:documentation> A reference to the name object that typifies this name. e.g. species for genus or genus for family etc. </xs:documentation> </xs:annotation> </xs:element> <xs:element name="LectotypePublication" type="ReferenceType" minOccurs="0"> <xs:annotation> <xs:documentation> A reference to the publication where the lectotypes were nominated. </xs:documentation> </xs:annotation> </xs:element> <xs:element name="LectotypeMicroReference" type="xs:string" minOccurs="0"> <xs:annotation> <xs:documentation> Specifies any minor reference parts connected to the lectotypification publication e.g. page number </xs:documentation> </xs:annotation> </xs:element> </xs:sequence> </xs:complexType> </xs:element> </xs:choice> </xs:sequence> </xs:complexType> </xs:element> <xs:element name="SpellingCorrectionOf" type="NomenclaturalNoteType" minOccurs="0" maxOccurs="unbounded"> <xs:annotation> <xs:documentation>The current name is a spelling correction of the name that is pointed to. </xs:documentation> </xs:annotation> </xs:element> <xs:element name="Basionym" type="NomenclaturalNoteType" minOccurs="0"> <xs:annotation> <xs:documentation> The current name is a recombination (comb nov) of the name pointed to and the name pointed to is not, itself, a recombination. </xs:documentation> </xs:annotation> </xs:element> <xs:element name="BasedOn" type="NomenclaturalNoteType" minOccurs="0"> <xs:annotation> <xs:documentation>The current name is the validation of a that was not fully published before. Covers the use of ex in botanical author strings. ICBN Art. 46.4: e.g. if this name object represents G. tomentosum Nutt. ex Seem. then the pointer should point to G. tomentosum Nutt. </xs:documentation> </xs:annotation> </xs:element> <xs:element name="ConservedAgainst" type="NomenclaturalNoteType" minOccurs="0" maxOccurs="unbounded"> <xs:annotation> <xs:documentation> The current name is conserved against the name pointed to. ICBN: Conservation is covered under Article 14 and Appendix II and Appendix III (this name is nomina conservanda). ICZN: Conservation is covered under Article 23.9 (this name is nomen protectum and the target name is nomen oblitum) </xs:documentation> </xs:annotation> </xs:element> <xs:element name="LaterHomonymOf" type="NomenclaturalNoteType" minOccurs="0"> <xs:annotation> <xs:documentation> Current name has same spelling as target name but was published later and has priority over it (unless conserved or sanctioned). See ICBN: Article 53, ICZN: Chapter 12, Article 52. </xs:documentation> </xs:annotation> </xs:element> <xs:element name="Sanctioned" type="NomenclaturalNoteType" minOccurs="0"> <xs:annotation> <xs:documentation> With reference to fungi. ICBN: Articles 13.1d and 15. ICZN: No equivalent term for animals</xs:documentation> </xs:annotation> </xs:element> <xs:element name="ReplacementNameFor" type="NomenclaturalNoteType" minOccurs="0"> <xs:annotation> <xs:documentation> Current name is replacement for target name. Also called 'Nomen Novum' or 'avowed substitute' ICBN: Article 7.3 ICZN: Article 60.3. </xs:documentation> </xs:annotation> </xs:element> <xs:element name="PublicationStatus" type="NomenclaturalNoteType" minOccurs="0"> <xs:annotation> <xs:documentation> Note concerning the publication status of the name. e.g. whether it is validly published. THIS SHOULD NOT BE USED TO IMPLY WHETHER THE NAME IS CONSIDERED TO REPRESENT AN ACCEPTED TAXON. An accepted taxon can only be represented as a TaxonConcept. </xs:documentation> </xs:annotation> </xs:element> <xs:element name="ProviderLink" type="xs:string" minOccurs="0"/> <xs:element name="ProviderSpecificData" type="PlaceholderType" minOccurs="0"> <xs:annotation> <xs:documentation>Mechanism to allow for the extension of the schema for specific applications.</xs:documentation> </xs:annotation> </xs:element> </xs:sequence> <xs:attribute name="id" type="xs:NMTOKEN" use="required"/> <xs:attribute name="isAnamorphic" type="xs:boolean" use="optional" default="false"> <xs:annotation> <xs:documentation> Some fungal names under ICBN are termed anamorphic. This is because they have asexual types and aren't available for use for teleomorphic or holomorphic taxa. Use this flag to indicate that the name is anamorphic. </xs:documentation> </xs:annotation> </xs:attribute> <xs:attribute name="nomenclaturalCode" type="NomenclaturalCodesEnum" use="required"> <xs:annotation> <xs:documentation> The nomenclatural code that governs the construction and use of this name. This is a TCS signature field. </xs:documentation> </xs:annotation> </xs:attribute> </xs:complexType> |
attribute ScientificName/@id
| type | xs:NMTOKEN | ||||
| properties |
|
||||
| source | <xs:attribute name="id" type="xs:NMTOKEN" use="required"/> |
attribute ScientificName/@isAnamorphic
| type | xs:boolean | ||||||
| properties |
|
||||||
| annotation |
|
||||||
| source | <xs:attribute name="isAnamorphic" type="xs:boolean" use="optional" default="false"> <xs:annotation> <xs:documentation> Some fungal names under ICBN are termed anamorphic. This is because they have asexual types and aren't available for use for teleomorphic or holomorphic taxa. Use this flag to indicate that the name is anamorphic. </xs:documentation> </xs:annotation> </xs:attribute> |
attribute ScientificName/@nomenclaturalCode
| type | NomenclaturalCodesEnum | |||||||||||||||||||||||||||||
| properties |
|
|||||||||||||||||||||||||||||
| facets |
|
|||||||||||||||||||||||||||||
| annotation |
|
|||||||||||||||||||||||||||||
| source | <xs:attribute name="nomenclaturalCode" type="NomenclaturalCodesEnum" use="required"> <xs:annotation> <xs:documentation> The nomenclatural code that governs the construction and use of this name. This is a TCS signature field. </xs:documentation> </xs:annotation> </xs:attribute> |
element ScientificName/Simple
| diagram |  |
||||
| namespace | http://www.tdwg.org/schemas/tcs/1.01 | ||||
| type | xs:string | ||||
| properties |
|
||||
| annotation |
|
||||
| source | <xs:element name="Simple" type="xs:string"> <xs:annotation> <xs:documentation> A full (non-atomized) name, including nomenclatural citation (authors/year in the form mandated by the relevant code). The name should be normalized (removing optional infrageneric or infraspecific hierarchy, author given only for the lowest nomenclatural rank) human readable and unique representation of the nomenclatoral name object. Where available, this should be generated from the canonical name and authorship plus Name extensions. In combination with the ID this constitutes the lowest abstraction a of name. Any other information is optional. </xs:documentation> </xs:annotation> </xs:element> |
element ScientificName/Rank
| diagram | 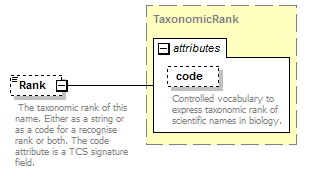 |
||||||||||||||
| namespace | http://www.tdwg.org/schemas/tcs/1.01 | ||||||||||||||
| type | TaxonomicRank | ||||||||||||||
| properties |
|
||||||||||||||
| attributes |
|
||||||||||||||
| annotation |
|
||||||||||||||
| source | <xs:element name="Rank" type="TaxonomicRank" minOccurs="0"> <xs:annotation> <xs:documentation> The taxonomic rank of this name. Either as a string or as a code for a recognise rank or both. The code attribute is a TCS signature field. </xs:documentation> </xs:annotation> </xs:element> |
element ScientificName/CanonicalName
| diagram | 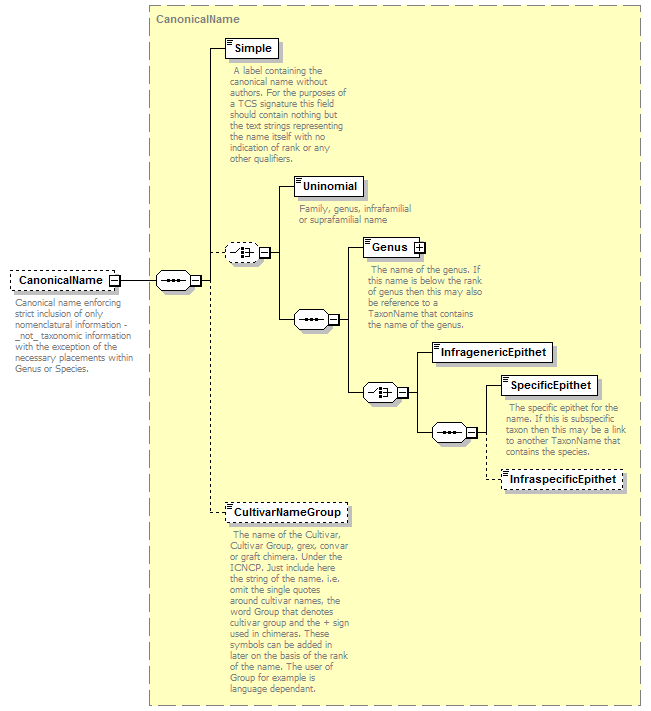 |
||||||||
| namespace | http://www.tdwg.org/schemas/tcs/1.01 | ||||||||
| type | CanonicalName | ||||||||
| properties |
|
||||||||
| children | Simple Uninomial Genus InfragenericEpithet SpecificEpithet InfraspecificEpithet CultivarNameGroup | ||||||||
| annotation |
|
||||||||
| source | <xs:element name="CanonicalName" type="CanonicalName" minOccurs="0"> <xs:annotation> <xs:documentation>Canonical name enforcing strict inclusion of only nomenclatural information - _not_ taxonomic information with the exception of the necessary placements within Genus or Species. </xs:documentation> </xs:annotation> </xs:element> |
element ScientificName/CanonicalAuthorship
| diagram | 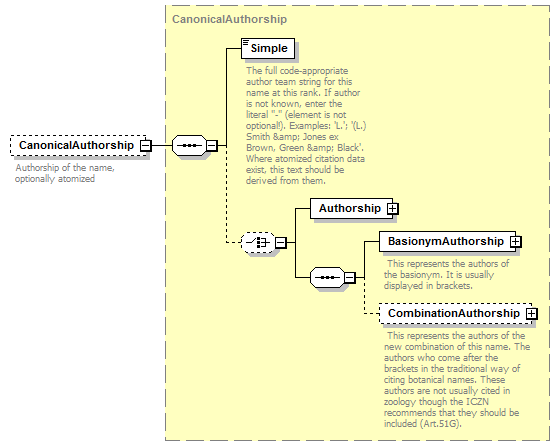 |
||||||||
| namespace | http://www.tdwg.org/schemas/tcs/1.01 | ||||||||
| type | CanonicalAuthorship | ||||||||
| properties |
|
||||||||
| children | Simple Authorship BasionymAuthorship CombinationAuthorship | ||||||||
| annotation |
|
||||||||
| source | <xs:element name="CanonicalAuthorship" type="CanonicalAuthorship" minOccurs="0"> <xs:annotation> <xs:documentation>Authorship of the name, optionally atomized</xs:documentation> </xs:annotation> </xs:element> |
element ScientificName/PublishedIn
| diagram | 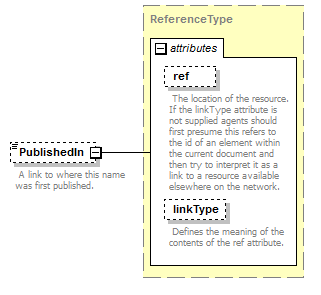 |
||||||||||||||||||||||
| namespace | http://www.tdwg.org/schemas/tcs/1.01 | ||||||||||||||||||||||
| type | ReferenceType | ||||||||||||||||||||||
| properties |
|
||||||||||||||||||||||
| attributes |
|
||||||||||||||||||||||
| annotation |
|
||||||||||||||||||||||
| source | <xs:element name="PublishedIn" type="ReferenceType" minOccurs="0"> <xs:annotation> <xs:documentation> A link to where this name was first published. </xs:documentation> </xs:annotation> </xs:element> |
element ScientificName/Year
| diagram |  |
||||||||
| namespace | http://www.tdwg.org/schemas/tcs/1.01 | ||||||||
| type | xs:gYear | ||||||||
| properties |
|
||||||||
| annotation |
|
||||||||
| source | <xs:element name="Year" type="xs:gYear" minOccurs="0"> <xs:annotation> <xs:documentation> The year of publication of the name as it would appear in the publication. This is the publication year for THIS name combination not for the basionym should this be a comb nov. This is a TCS signature field. </xs:documentation> </xs:annotation> </xs:element> |
element ScientificName/MicroReference
| diagram |  |
||||||||
| namespace | http://www.tdwg.org/schemas/tcs/1.01 | ||||||||
| type | xs:string | ||||||||
| properties |
|
||||||||
| annotation |
|
||||||||
| source | <xs:element name="MicroReference" type="xs:string" minOccurs="0"> <xs:annotation> <xs:documentation> Specifies any minor reference parts connected to the name publication e.g. page number </xs:documentation> </xs:annotation> </xs:element> |
element ScientificName/Typification
| diagram |  |
||||||||
| namespace | http://www.tdwg.org/schemas/tcs/1.01 | ||||||||
| properties |
|
||||||||
| children | Simple TypeVouchers TypeName | ||||||||
| source | <xs:element name="Typification" minOccurs="0"> <xs:complexType> <xs:sequence> <xs:element name="Simple" type="xs:string"/> <xs:choice> <xs:element name="TypeVouchers"> <xs:annotation> <xs:documentation> A container for type specimens and other vouchers. </xs:documentation> </xs:annotation> <xs:complexType> <xs:sequence> <xs:element name="TypeVoucher" maxOccurs="unbounded"> <xs:annotation> <xs:documentation>Names at species and below are typified by specimens.</xs:documentation> </xs:annotation> <xs:complexType> <xs:sequence> <xs:element name="VoucherReference" type="ReferenceType" minOccurs="0"> <xs:annotation> <xs:documentation> A reference to a type specimen for this name. </xs:documentation> </xs:annotation> </xs:element> <xs:element name="LectotypePublication" type="ReferenceType" minOccurs="0"> <xs:annotation> <xs:documentation> A reference to the publication where the lectotype was nominated. </xs:documentation> </xs:annotation> </xs:element> <xs:element name="LectotypeMicroReference" type="xs:string" minOccurs="0"> <xs:annotation> <xs:documentation> Specifies any minor reference parts connected to the lectotypification publication e.g. page number </xs:documentation> </xs:annotation> </xs:element> </xs:sequence> <xs:attribute name="typeOfType" type="NomenclaturalTypeStatusOfUnitsEnum"> <xs:annotation> <xs:documentation> The kind of type this specimen is e.g. paratype, isotype, holotype etc. </xs:documentation> </xs:annotation> </xs:attribute> </xs:complexType> </xs:element> </xs:sequence> </xs:complexType> </xs:element> <xs:element name="TypeName"> <xs:annotation> <xs:documentation>Names above species level are typified by the NAME of a lower taxon.</xs:documentation> </xs:annotation> <xs:complexType> <xs:sequence> <xs:element name="NameReference" type="ReferenceType" minOccurs="0"> <xs:annotation> <xs:documentation> A reference to the name object that typifies this name. e.g. species for genus or genus for family etc. </xs:documentation> </xs:annotation> </xs:element> <xs:element name="LectotypePublication" type="ReferenceType" minOccurs="0"> <xs:annotation> <xs:documentation> A reference to the publication where the lectotypes were nominated. </xs:documentation> </xs:annotation> </xs:element> <xs:element name="LectotypeMicroReference" type="xs:string" minOccurs="0"> <xs:annotation> <xs:documentation> Specifies any minor reference parts connected to the lectotypification publication e.g. page number </xs:documentation> </xs:annotation> </xs:element> </xs:sequence> </xs:complexType> </xs:element> </xs:choice> </xs:sequence> </xs:complexType> </xs:element> |
element ScientificName/Typification/Simple
| diagram |  |
||||
| namespace | http://www.tdwg.org/schemas/tcs/1.01 | ||||
| type | xs:string | ||||
| properties |
|
||||
| source | <xs:element name="Simple" type="xs:string"/> |
element ScientificName/Typification/TypeVouchers
| diagram |  |
||||
| namespace | http://www.tdwg.org/schemas/tcs/1.01 | ||||
| properties |
|
||||
| children | TypeVoucher | ||||
| annotation |
|
||||
| source | <xs:element name="TypeVouchers"> <xs:annotation> <xs:documentation> A container for type specimens and other vouchers. </xs:documentation> </xs:annotation> <xs:complexType> <xs:sequence> <xs:element name="TypeVoucher" maxOccurs="unbounded"> <xs:annotation> <xs:documentation>Names at species and below are typified by specimens.</xs:documentation> </xs:annotation> <xs:complexType> <xs:sequence> <xs:element name="VoucherReference" type="ReferenceType" minOccurs="0"> <xs:annotation> <xs:documentation> A reference to a type specimen for this name. </xs:documentation> </xs:annotation> </xs:element> <xs:element name="LectotypePublication" type="ReferenceType" minOccurs="0"> <xs:annotation> <xs:documentation> A reference to the publication where the lectotype was nominated. </xs:documentation> </xs:annotation> </xs:element> <xs:element name="LectotypeMicroReference" type="xs:string" minOccurs="0"> <xs:annotation> <xs:documentation> Specifies any minor reference parts connected to the lectotypification publication e.g. page number </xs:documentation> </xs:annotation> </xs:element> </xs:sequence> <xs:attribute name="typeOfType" type="NomenclaturalTypeStatusOfUnitsEnum"> <xs:annotation> <xs:documentation> The kind of type this specimen is e.g. paratype, isotype, holotype etc. </xs:documentation> </xs:annotation> </xs:attribute> </xs:complexType> </xs:element> </xs:sequence> </xs:complexType> </xs:element> |
element ScientificName/Typification/TypeVouchers/TypeVoucher
| diagram | 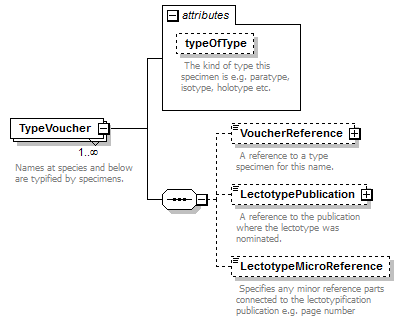 |
||||||||||||||
| namespace | http://www.tdwg.org/schemas/tcs/1.01 | ||||||||||||||
| properties |
|
||||||||||||||
| children | VoucherReference LectotypePublication LectotypeMicroReference | ||||||||||||||
| attributes |
|
||||||||||||||
| annotation |
|
||||||||||||||
| source | <xs:element name="TypeVoucher" maxOccurs="unbounded"> <xs:annotation> <xs:documentation>Names at species and below are typified by specimens.</xs:documentation> </xs:annotation> <xs:complexType> <xs:sequence> <xs:element name="VoucherReference" type="ReferenceType" minOccurs="0"> <xs:annotation> <xs:documentation> A reference to a type specimen for this name. </xs:documentation> </xs:annotation> </xs:element> <xs:element name="LectotypePublication" type="ReferenceType" minOccurs="0"> <xs:annotation> <xs:documentation> A reference to the publication where the lectotype was nominated. </xs:documentation> </xs:annotation> </xs:element> <xs:element name="LectotypeMicroReference" type="xs:string" minOccurs="0"> <xs:annotation> <xs:documentation> Specifies any minor reference parts connected to the lectotypification publication e.g. page number </xs:documentation> </xs:annotation> </xs:element> </xs:sequence> <xs:attribute name="typeOfType" type="NomenclaturalTypeStatusOfUnitsEnum"> <xs:annotation> <xs:documentation> The kind of type this specimen is e.g. paratype, isotype, holotype etc. </xs:documentation> </xs:annotation> </xs:attribute> </xs:complexType> </xs:element> |
attribute ScientificName/Typification/TypeVouchers/TypeVoucher/@typeOfType
| type | NomenclaturalTypeStatusOfUnitsEnum | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| properties |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| facets |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| annotation |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| source | <xs:attribute name="typeOfType" type="NomenclaturalTypeStatusOfUnitsEnum"> <xs:annotation> <xs:documentation> The kind of type this specimen is e.g. paratype, isotype, holotype etc. </xs:documentation> </xs:annotation> </xs:attribute> |
element ScientificName/Typification/TypeVouchers/TypeVoucher/VoucherReference
| diagram |  |
||||||||||||||||||||||
| namespace | http://www.tdwg.org/schemas/tcs/1.01 | ||||||||||||||||||||||
| type | ReferenceType | ||||||||||||||||||||||
| properties |
|
||||||||||||||||||||||
| attributes |
|
||||||||||||||||||||||
| annotation |
|
||||||||||||||||||||||
| source | <xs:element name="VoucherReference" type="ReferenceType" minOccurs="0"> <xs:annotation> <xs:documentation> A reference to a type specimen for this name. </xs:documentation> </xs:annotation> </xs:element> |
element ScientificName/Typification/TypeVouchers/TypeVoucher/LectotypePublication
| diagram |  |
||||||||||||||||||||||
| namespace | http://www.tdwg.org/schemas/tcs/1.01 | ||||||||||||||||||||||
| type | ReferenceType | ||||||||||||||||||||||
| properties |
|
||||||||||||||||||||||
| attributes |
|
||||||||||||||||||||||
| annotation |
|
||||||||||||||||||||||
| source | <xs:element name="LectotypePublication" type="ReferenceType" minOccurs="0"> <xs:annotation> <xs:documentation> A reference to the publication where the lectotype was nominated. </xs:documentation> </xs:annotation> </xs:element> |
element ScientificName/Typification/TypeVouchers/TypeVoucher/LectotypeMicroReference
| diagram |  |
||||||||
| namespace | http://www.tdwg.org/schemas/tcs/1.01 | ||||||||
| type | xs:string | ||||||||
| properties |
|
||||||||
| annotation |
|
||||||||
| source | <xs:element name="LectotypeMicroReference" type="xs:string" minOccurs="0"> <xs:annotation> <xs:documentation> Specifies any minor reference parts connected to the lectotypification publication e.g. page number </xs:documentation> </xs:annotation> </xs:element> |
element ScientificName/Typification/TypeName
| diagram |  |
||||
| namespace | http://www.tdwg.org/schemas/tcs/1.01 | ||||
| properties |
|
||||
| children | NameReference LectotypePublication LectotypeMicroReference | ||||
| annotation |
|
||||
| source | <xs:element name="TypeName"> <xs:annotation> <xs:documentation>Names above species level are typified by the NAME of a lower taxon.</xs:documentation> </xs:annotation> <xs:complexType> <xs:sequence> <xs:element name="NameReference" type="ReferenceType" minOccurs="0"> <xs:annotation> <xs:documentation> A reference to the name object that typifies this name. e.g. species for genus or genus for family etc. </xs:documentation> </xs:annotation> </xs:element> <xs:element name="LectotypePublication" type="ReferenceType" minOccurs="0"> <xs:annotation> <xs:documentation> A reference to the publication where the lectotypes were nominated. </xs:documentation> </xs:annotation> </xs:element> <xs:element name="LectotypeMicroReference" type="xs:string" minOccurs="0"> <xs:annotation> <xs:documentation> Specifies any minor reference parts connected to the lectotypification publication e.g. page number </xs:documentation> </xs:annotation> </xs:element> </xs:sequence> </xs:complexType> </xs:element> |
element ScientificName/Typification/TypeName/NameReference
| diagram |  |
||||||||||||||||||||||
| namespace | http://www.tdwg.org/schemas/tcs/1.01 | ||||||||||||||||||||||
| type | ReferenceType | ||||||||||||||||||||||
| properties |
|
||||||||||||||||||||||
| attributes |
|
||||||||||||||||||||||
| annotation |
|
||||||||||||||||||||||
| source | <xs:element name="NameReference" type="ReferenceType" minOccurs="0"> <xs:annotation> <xs:documentation> A reference to the name object that typifies this name. e.g. species for genus or genus for family etc. </xs:documentation> </xs:annotation> </xs:element> |
element ScientificName/Typification/TypeName/LectotypePublication
| diagram |  |
||||||||||||||||||||||
| namespace | http://www.tdwg.org/schemas/tcs/1.01 | ||||||||||||||||||||||
| type | ReferenceType | ||||||||||||||||||||||
| properties |
|
||||||||||||||||||||||
| attributes |
|
||||||||||||||||||||||
| annotation |
|
||||||||||||||||||||||
| source | <xs:element name="LectotypePublication" type="ReferenceType" minOccurs="0"> <xs:annotation> <xs:documentation> A reference to the publication where the lectotypes were nominated. </xs:documentation> </xs:annotation> </xs:element> |
element ScientificName/Typification/TypeName/LectotypeMicroReference
| diagram |  |
||||||||
| namespace | http://www.tdwg.org/schemas/tcs/1.01 | ||||||||
| type | xs:string | ||||||||
| properties |
|
||||||||
| annotation |
|
||||||||
| source | <xs:element name="LectotypeMicroReference" type="xs:string" minOccurs="0"> <xs:annotation> <xs:documentation> Specifies any minor reference parts connected to the lectotypification publication e.g. page number </xs:documentation> </xs:annotation> </xs:element> |
element ScientificName/SpellingCorrectionOf
| diagram | 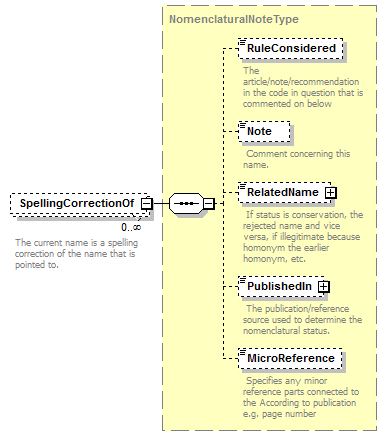 |
||||||||
| namespace | http://www.tdwg.org/schemas/tcs/1.01 | ||||||||
| type | NomenclaturalNoteType | ||||||||
| properties |
|
||||||||
| children | RuleConsidered Note RelatedName PublishedIn MicroReference | ||||||||
| annotation |
|
||||||||
| source | <xs:element name="SpellingCorrectionOf" type="NomenclaturalNoteType" minOccurs="0" maxOccurs="unbounded"> <xs:annotation> <xs:documentation>The current name is a spelling correction of the name that is pointed to. </xs:documentation> </xs:annotation> </xs:element> |
element ScientificName/Basionym
| diagram | 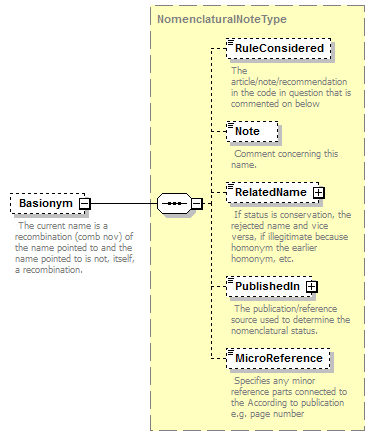 |
||||||||
| namespace | http://www.tdwg.org/schemas/tcs/1.01 | ||||||||
| type | NomenclaturalNoteType | ||||||||
| properties |
|
||||||||
| children | RuleConsidered Note RelatedName PublishedIn MicroReference | ||||||||
| annotation |
|
||||||||
| source | <xs:element name="Basionym" type="NomenclaturalNoteType" minOccurs="0"> <xs:annotation> <xs:documentation> The current name is a recombination (comb nov) of the name pointed to and the name pointed to is not, itself, a recombination. </xs:documentation> </xs:annotation> </xs:element> |
element ScientificName/BasedOn
| diagram | 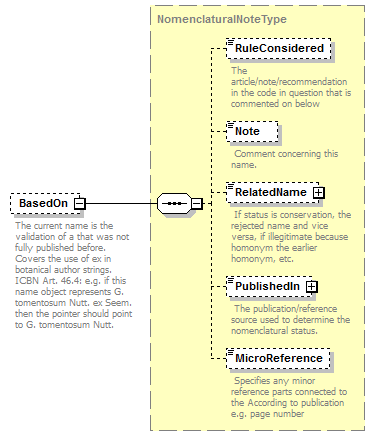 |
||||||||
| namespace | http://www.tdwg.org/schemas/tcs/1.01 | ||||||||
| type | NomenclaturalNoteType | ||||||||
| properties |
|
||||||||
| children | RuleConsidered Note RelatedName PublishedIn MicroReference | ||||||||
| annotation |
|
||||||||
| source | <xs:element name="BasedOn" type="NomenclaturalNoteType" minOccurs="0"> <xs:annotation> <xs:documentation>The current name is the validation of a that was not fully published before. Covers the use of ex in botanical author strings. ICBN Art. 46.4: e.g. if this name object represents G. tomentosum Nutt. ex Seem. then the pointer should point to G. tomentosum Nutt. </xs:documentation> </xs:annotation> </xs:element> |
element ScientificName/ConservedAgainst
| diagram | 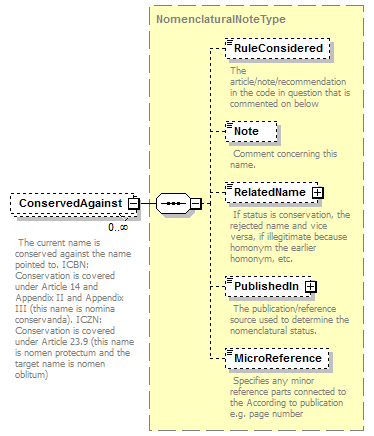 |
||||||||
| namespace | http://www.tdwg.org/schemas/tcs/1.01 | ||||||||
| type | NomenclaturalNoteType | ||||||||
| properties |
|
||||||||
| children | RuleConsidered Note RelatedName PublishedIn MicroReference | ||||||||
| annotation |
|
||||||||
| source | <xs:element name="ConservedAgainst" type="NomenclaturalNoteType" minOccurs="0" maxOccurs="unbounded"> <xs:annotation> <xs:documentation> The current name is conserved against the name pointed to. ICBN: Conservation is covered under Article 14 and Appendix II and Appendix III (this name is nomina conservanda). ICZN: Conservation is covered under Article 23.9 (this name is nomen protectum and the target name is nomen oblitum) </xs:documentation> </xs:annotation> </xs:element> |
element ScientificName/LaterHomonymOf
| diagram | 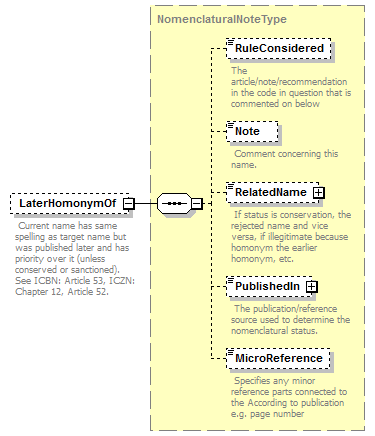 |
||||||||
| namespace | http://www.tdwg.org/schemas/tcs/1.01 | ||||||||
| type | NomenclaturalNoteType | ||||||||
| properties |
|
||||||||
| children | RuleConsidered Note RelatedName PublishedIn MicroReference | ||||||||
| annotation |
|
||||||||
| source | <xs:element name="LaterHomonymOf" type="NomenclaturalNoteType" minOccurs="0"> <xs:annotation> <xs:documentation> Current name has same spelling as target name but was published later and has priority over it (unless conserved or sanctioned). See ICBN: Article 53, ICZN: Chapter 12, Article 52. </xs:documentation> </xs:annotation> </xs:element> |
element ScientificName/Sanctioned
| diagram | 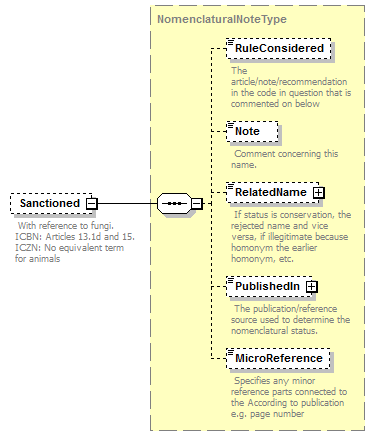 |
||||||||
| namespace | http://www.tdwg.org/schemas/tcs/1.01 | ||||||||
| type | NomenclaturalNoteType | ||||||||
| properties |
|
||||||||
| children | RuleConsidered Note RelatedName PublishedIn MicroReference | ||||||||
| annotation |
|
||||||||
| source | <xs:element name="Sanctioned" type="NomenclaturalNoteType" minOccurs="0"> <xs:annotation> <xs:documentation> With reference to fungi. ICBN: Articles 13.1d and 15. ICZN: No equivalent term for animals</xs:documentation> </xs:annotation> </xs:element> |
element ScientificName/ReplacementNameFor
| diagram | 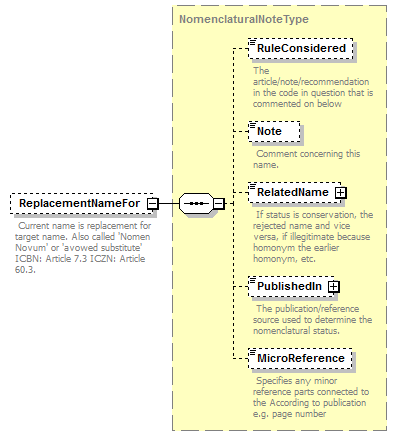 |
||||||||
| namespace | http://www.tdwg.org/schemas/tcs/1.01 | ||||||||
| type | NomenclaturalNoteType | ||||||||
| properties |
|
||||||||
| children | RuleConsidered Note RelatedName PublishedIn MicroReference | ||||||||
| annotation |
|
||||||||
| source | <xs:element name="ReplacementNameFor" type="NomenclaturalNoteType" minOccurs="0"> <xs:annotation> <xs:documentation> Current name is replacement for target name. Also called 'Nomen Novum' or 'avowed substitute' ICBN: Article 7.3 ICZN: Article 60.3. </xs:documentation> </xs:annotation> </xs:element> |
element ScientificName/PublicationStatus
| diagram | 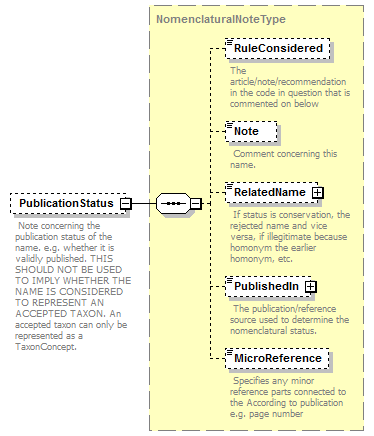 |
||||||||
| namespace | http://www.tdwg.org/schemas/tcs/1.01 | ||||||||
| type | NomenclaturalNoteType | ||||||||
| properties |
|
||||||||
| children | RuleConsidered Note RelatedName PublishedIn MicroReference | ||||||||
| annotation |
|
||||||||
| source | <xs:element name="PublicationStatus" type="NomenclaturalNoteType" minOccurs="0"> <xs:annotation> <xs:documentation> Note concerning the publication status of the name. e.g. whether it is validly published. THIS SHOULD NOT BE USED TO IMPLY WHETHER THE NAME IS CONSIDERED TO REPRESENT AN ACCEPTED TAXON. An accepted taxon can only be represented as a TaxonConcept. </xs:documentation> </xs:annotation> </xs:element> |
element ScientificName/ProviderLink
| diagram |  |
||||||||
| namespace | http://www.tdwg.org/schemas/tcs/1.01 | ||||||||
| type | xs:string | ||||||||
| properties |
|
||||||||
| source | <xs:element name="ProviderLink" type="xs:string" minOccurs="0"/> |
element ScientificName/ProviderSpecificData
| diagram |  |
||||||||
| namespace | http://www.tdwg.org/schemas/tcs/1.01 | ||||||||
| type | PlaceholderType | ||||||||
| properties |
|
||||||||
| attributes |
|
||||||||
| annotation |
|
||||||||
| source | <xs:element name="ProviderSpecificData" type="PlaceholderType" minOccurs="0"> <xs:annotation> <xs:documentation>Mechanism to allow for the extension of the schema for specific applications.</xs:documentation> </xs:annotation> </xs:element> |
complexType TaxonomicRank
| diagram |  |
||||||||||||||
| namespace | http://www.tdwg.org/schemas/tcs/1.01 | ||||||||||||||
| properties |
|
||||||||||||||
| used by |
|
||||||||||||||
| attributes |
|
||||||||||||||
| annotation |
|
||||||||||||||
| source | <xs:complexType name="TaxonomicRank" mixed="true"> <xs:annotation> <xs:documentation>Taxonomic ranks can be represented as either a plain text string in the body of the element or an attribute from a controlled vocabulary. It is highly recommended that the attribute is used where ever possible. </xs:documentation> </xs:annotation> <xs:attribute name="code" type="TaxonomicRankEnum" use="optional"> <xs:annotation> <xs:documentation>Controlled vocabulary to express taxonomic rank of scientific names in biology.</xs:documentation> </xs:annotation> </xs:attribute> </xs:complexType> |
attribute TaxonomicRank/@code
| type | TaxonomicRankEnum | ||||
| properties |
|
||||
| annotation |
|
||||
| source | <xs:attribute name="code" type="TaxonomicRankEnum" use="optional"> <xs:annotation> <xs:documentation>Controlled vocabulary to express taxonomic rank of scientific names in biology.</xs:documentation> </xs:annotation> </xs:attribute> |
simpleType NomenclaturalCodesEnum
| namespace | http://www.tdwg.org/schemas/tcs/1.01 | |||||||||||||||||||||||||||||
| type | restriction of xs:Name | |||||||||||||||||||||||||||||
| properties |
|
|||||||||||||||||||||||||||||
| used by |
|
|||||||||||||||||||||||||||||
| facets |
|
|||||||||||||||||||||||||||||
| annotation |
|
|||||||||||||||||||||||||||||
| source | <xs:simpleType name="NomenclaturalCodesEnum"> <xs:annotation> <xs:documentation>Enumeration of the nomenclatural code under which a name is considered valid. (Source: comparison of enumerations in ABCD 1.49 and initial LinneanCore.)</xs:documentation> </xs:annotation> <xs:restriction base="xs:Name"> <xs:enumeration value="Viral"/> <xs:enumeration value="Bacteriological"/> <xs:enumeration value="Botanical"> <xs:annotation> <xs:documentation>International Code of Botanical Nomenclature, ICBN</xs:documentation> </xs:annotation> </xs:enumeration> <xs:enumeration value="Zoological"> <xs:annotation> <xs:documentation>International Code of Zoological Nomenclature, ICZN</xs:documentation> </xs:annotation> </xs:enumeration> <xs:enumeration value="CultivatedPlant"> <xs:annotation> <xs:documentation>International Code of cultivated plants, ICNCP</xs:documentation> </xs:annotation> </xs:enumeration> <xs:enumeration value="Indeterminate"> <xs:annotation> <xs:documentation>Examples: ambiregnally named taxa can not be fixed to a code; to express the (pre-starting point) names on which the sanctioning mechanism of fungal names is based.</xs:documentation> </xs:annotation> </xs:enumeration> </xs:restriction> </xs:simpleType> |
simpleType NomenclaturalTypeStatusOfUnitsEnum
| namespace | http://www.tdwg.org/schemas/tcs/1.01 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| type | restriction of xs:Name | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| properties |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| used by |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| facets |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| annotation |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| source | <xs:simpleType name="NomenclaturalTypeStatusOfUnitsEnum"> <xs:annotation> <xs:documentation> <p>This list is a first version of a constrained vocabulary to express typifying relations between taxonomic names and units (specimens or objects preserved in collections). Beyond those type categories explicitly governed by nomenclatural codes (Zoology, Botany, Bacterioloy, Virology), the list also includes some additional type status terms. These categories may be helpful when interpreting the original circumscription (topotypes, ex-types), but do not have the same binding status as terms governed by the nomenclatural codes. The enumeration attempts to strike a balance between listing all possible terms, and remaining comprehensible. In general, including too many terms was considered less problematic than omitting terms. Applications may easily select a subset for presentation in their user interface.</p> <p>This list is intended as a first version and it is hoped that in the review process through TDWG it will achieve sufficient maturity to be truly useful. It is expected that over time revisions will have to be made. Please use the WIKI (http://efgblade.cs.umb.edu/twiki/bin/view/UBIF/NomenclaturalTypeStatusOfUnitsDiscussion) to discuss the current list and the lists of synonymous, doubtful, or excluded type terms provided therein.</p> <p>Some background information: A type provides the objective standard of reference to determine the application of a taxon name. The type status of a unit (specimen) is only meaningful in combination with the name that is being typified (a unit may have been designated type for multiple names in different publications). The type status of an object may be designated in the original description of a scientific name (original designation), or - under rules layed out in the respective nomenclatural codes - at a later time (subsequent designation). -- For taxa above species rank the type is always a lower rank taxon (e. g., species for genus, genus for family). The type terms for this situation are not included in the enumeration. Ultimately, typication of all taxa goes back to physical type units, but this should not be recorded as such in data sets. The indirect type reference in higher taxa means that typification changes to the lower taxon automatically affect the higher taxon.</p> <p>The exact definitions of type status differ between nomenclatural codes (ICBN, ICZN, ICNP/ICNB, etc.). The term definitions are intended to be informative and generally applicable across the different codes. The should not be interpreted as authoritative; in nomenclatural work the exact definitions in the respective codes have to be consulted. A duplication of status codes (bot-holo, zoo-holo, bact-holo, etc.) is not considered desirable or necessary. Since the application of the type status terms is constrained by the relationship of the typified name with a specific code, the exact definition can always be unambiguously retrieved.</p> <p>The following publications have been consulted to determine the number of type terms that should be included and to prepare the semantic definitions:</p> <ul> <li>Nomenclatural Glossary for Zoology (January 18 2000; ftp://ftp.york.biosis.org/sysgloss.txt; verified 17. June 2004)</li> <li>ICBN St. Louis Code (http://www.bgbm.fu-berlin.de/iapt/nomenclature/code/SaintLouis/0013Ch2Sec2a009.htm; verified 17. June 2004)</li> <li>Draft BioCode 4th version (Greuter et al., 1997; http://www.rom.on.ca/biodiversity/biocode/biocode1997.html)</li> <li>Glossary of 'type' terminology (Ronald H. Petersen; http://fp.bio.utk.edu/mycology/Nomenclature/nom-type.htm)</li> <li>Dictionary of Ichthyology (Brian W. Coad and Don E. McAllister, 2004; http://www.briancoad.com/Dictionary/introduction.htm)</li> <li>A useful resource that was not available when writing this proposal might be: Hawksworth, D.L., W.G. Chaloner, O. Krauss, J. McNeill, M.A. Mayo, D.H. Nicolson, P.H.A. Sneath, R.P. Trehane and P.K. Tubbs. 1994. A draft Glossary of terms used in Bionomenclature. (IUBS Monogr. 9) International Union of Biological Sciences, Paris. 74 pp.</li> </ul> <p>Many thanks for review and help to Dr. Miguel A. Alonso-Zarazaga and Dr. Walter Gams. Gregor Hagedorn, 13.7.2004</p> </xs:documentation> </xs:annotation> <xs:restriction base="xs:Name"> <xs:enumeration value="allo"> <xs:annotation> <xs:documentation>Allotype -- A paratype specimen designated from the type series by the original author that is the opposite sex of the holotype. The term is not regulated by the ICZN. [Zoo.]</xs:documentation> </xs:annotation> </xs:enumeration> <xs:enumeration value="allolecto"> <xs:annotation> <xs:documentation>Allolectotype -- A paralectotype specimen that is the opposite sex of the lectotype. The term is not regulated by the ICZN. [Zoo.]</xs:documentation> </xs:annotation> </xs:enumeration> <xs:enumeration value="alloneo"> <xs:annotation> <xs:documentation>Alloneotype -- A paraneotype specimen that is the opposite sex of the neotype. The term is not regulated by the ICZN. [Zoo.]</xs:documentation> </xs:annotation> </xs:enumeration> <xs:enumeration value="co"> <xs:annotation> <xs:documentation>Cotype -- A deprecated term no longer recognized in the ICZN; formerly used for either syntype or paratype [see ICZN Recommendation 73E]. [Zoo.]</xs:documentation> </xs:annotation> </xs:enumeration> <xs:enumeration value="epi"> <xs:annotation> <xs:documentation>Epitype -- An epitype is a specimen or illustration selected to serve as an interpretative type when any kind of holotype, lectotype, etc. is demonstrably ambiguous and cannot be critically identified for purposes of the precise application of the name of a taxon (see Art. ICBN 9.7, 9.18). An epitype supplements, rather than replaces existing types. [Bot./Bio.]</xs:documentation> </xs:annotation> </xs:enumeration> <xs:enumeration value="ex"> <xs:annotation> <xs:documentation>Ex-Type -- A strain or cultivation derived from some kind of type material. Ex-types are not regulated by the botanical or zoological code. [Zoo./Bot.]</xs:documentation> </xs:annotation> </xs:enumeration> <xs:enumeration value="exepi"> <xs:annotation> <xs:documentation>Ex-Epitype -- A strain or cultivation derived from epitype material. Ex-types are not regulated by the botanical or zoological code. [Bot.]</xs:documentation> </xs:annotation> </xs:enumeration> <xs:enumeration value="exholo"> <xs:annotation> <xs:documentation>Ex-Holotype -- A strain or cultivation derived from holotype material. Ex-types are not regulated by the botanical or zoological code. [Zoo./Bot.]</xs:documentation> </xs:annotation> </xs:enumeration> <xs:enumeration value="exiso"> <xs:annotation> <xs:documentation>Ex-Isotype -- A strain or cultivation derived from isotype material. Ex-types are not regulated by the botanical or zoological code. [Zoo./Bot.]</xs:documentation> </xs:annotation> </xs:enumeration> <xs:enumeration value="exlecto"> <xs:annotation> <xs:documentation>Ex-Lectotype -- A strain or cultivation derived from lectotype material. Ex-types are not regulated by the botanical or zoological code. [Zoo./Bot.]</xs:documentation> </xs:annotation> </xs:enumeration> <xs:enumeration value="exneo"> <xs:annotation> <xs:documentation>Ex-Neotype -- A strain or cultivation derived from neotype material. Ex-types are not regulated by the botanical or zoological code. [Zoo./Bot.]</xs:documentation> </xs:annotation> </xs:enumeration> <xs:enumeration value="expara"> <xs:annotation> <xs:documentation>Ex-Paratype -- A strain or cultivation derived from paratype material. Ex-types are not regulated by the botanical or zoological code. [Zoo./Bot.]</xs:documentation> </xs:annotation> </xs:enumeration> <xs:enumeration value="exsyn"> <xs:annotation> <xs:documentation>Ex-Syntype -- A strain or cultivation derived from neotype material. Ex-types are not regulated by the botanical or zoological code. [Zoo./Bot.]</xs:documentation> </xs:annotation> </xs:enumeration> <xs:enumeration value="hapanto"> <xs:annotation> <xs:documentation>Hapantotype -- One or more preparations of directly related individuals representing distinct stages in the life cycle, which together form the type in an extant species of protistan [ICZN Article 72.5.4]. A hapantotype, while a series of individuals, is a holotype that must not be restricted by lectotype selection. If a hapantotype is found to contain individuals of more than one species, however, components may be excluded until it contains individuals of only one species [ICZN Article 73.3.2]. [Zoo.]</xs:documentation> </xs:annotation> </xs:enumeration> <xs:enumeration value="holo"> <xs:annotation> <xs:documentation>Holotype -- The one specimen or other element used or designated by the original author at the time of publication of the original description as the nomenclatural type of a species or infraspecific taxon. A holotype may be 'explicit' if it is clearly stated in the originating publication or 'implicit' if it is the single specimen proved to have been in the hands of the originating author when the description was published. [Zoo./Bot./Bio.]</xs:documentation> </xs:annotation> </xs:enumeration> <xs:enumeration value="icono"> <xs:annotation> <xs:documentation>Iconotype -- A drawing or photograph (also called 'phototype') of a type specimen. Note: the term "iconotype" is not used in the ICBN, but implicit in, e. g., ICBN Art. 7 and 38. [Zoo./Bot.]</xs:documentation> </xs:annotation> </xs:enumeration> <xs:enumeration value="iso"> <xs:annotation> <xs:documentation>Isotype -- An isotype is any duplicate of the holotype (i. e. part of a single gathering made by a collector at one time, from which the holotype was derived); it is always a specimen (ICBN Art. 7). [Bot.]</xs:documentation> </xs:annotation> </xs:enumeration> <xs:enumeration value="isolecto"> <xs:annotation> <xs:documentation>Isolectotype -- A duplicate of a neotype, compare lectotype. [Bot.]</xs:documentation> </xs:annotation> </xs:enumeration> <xs:enumeration value="isoneo"> <xs:annotation> <xs:documentation>Isoneotype -- A duplicate of a neotype, compare neotype. [Bot.]</xs:documentation> </xs:annotation> </xs:enumeration> <xs:enumeration value="isosyn"> <xs:annotation> <xs:documentation>Isosyntype -- A duplicate of a syntype, compare isotype = duplicate of holotype. [Bot.]</xs:documentation> </xs:annotation> </xs:enumeration> <xs:enumeration value="lecto"> <xs:annotation> <xs:documentation>Lectotype -- A specimen or other element designated subsequent to the publication of the original description from the original material (syntypes or paratypes) to serve as nomenclatural type. Lectotype designation can occur only where no holotype was designated at the time of publication or if it is missing (ICBN Art. 7, ICZN Art. 74). [Zoo./Bot.] -- Note: the BioCode defines lectotype as selection from holotype material in cases where the holotype material contains more than one taxon [Bio.].</xs:documentation> </xs:annotation> </xs:enumeration> <xs:enumeration value="neo"> <xs:annotation> <xs:documentation>Neotype -- A specimen designated as nomenclatural type subsequent to the publication of the original description in cases where the original holotype, lectotype, all paratypes and syntypes are lost or destroyed, or suppressed by the (botanical or zoological) commission on nomenclature. In zoology also called "Standard specimen" or "Representative specimen". [Zoo./Bot./Bio.]</xs:documentation> </xs:annotation> </xs:enumeration> <xs:enumeration value="para"> <xs:annotation> <xs:documentation>Paratype -- All of the specimens in the type series of a species or infraspecific taxon other than the holotype (and, in botany, isotypes). Paratypes must have been at the disposition of the author at the time when the original description was created and must have been designated and indicated in the publication. Judgment must be exercised on paratype status, for only rarely are specimens explicitly cited as paratypes, but usually as "specimens examined," "other material seen", etc. [Zoo./Bot.]</xs:documentation> </xs:annotation> </xs:enumeration> <xs:enumeration value="paralecto"> <xs:annotation> <xs:documentation>Paralectotype -- All of the specimens in the syntype series of a species or infraspecific taxon other than the lectotype itself. Also called "lectoparatype". [Zoo.]</xs:documentation> </xs:annotation> </xs:enumeration> <xs:enumeration value="paraneo"> <xs:annotation> <xs:documentation>Paraneotype -- All of the specimens in the syntype series of a species or infraspecific taxon other than the neotype itself. Also called "neoparatype". [Zoo.]</xs:documentation> </xs:annotation> </xs:enumeration> <xs:enumeration value="plasto"> <xs:annotation> <xs:documentation>Plastotype -- A copy or cast of type material, esp. relevant for fossil types. Not regulated by the botanical or zoological code (?). [Zoo./Bot.]</xs:documentation> </xs:annotation> </xs:enumeration> <xs:enumeration value="plastoholo"> <xs:annotation> <xs:documentation>Plastoholotype -- A copy or cast of holotype material (compare Plastotype).</xs:documentation> </xs:annotation> </xs:enumeration> <xs:enumeration value="plastoiso"> <xs:annotation> <xs:documentation>Plastoisotype -- A copy or cast of isotype material (compare Plastotype).</xs:documentation> </xs:annotation> </xs:enumeration> <xs:enumeration value="plastolecto"> <xs:annotation> <xs:documentation>Plastolectotype -- A copy or cast of lectotype material (compare Plastotype).</xs:documentation> </xs:annotation> </xs:enumeration> <xs:enumeration value="plastoneo"> <xs:annotation> <xs:documentation>Plastoneotype -- A copy or cast of neotype material (compare Plastotype).</xs:documentation> </xs:annotation> </xs:enumeration> <xs:enumeration value="plastopara"> <xs:annotation> <xs:documentation>Plastoparatype -- A copy or cast of paratype material (compare Plastotype).</xs:documentation> </xs:annotation> </xs:enumeration> <xs:enumeration value="plastosyn"> <xs:annotation> <xs:documentation>Plastosyntype -- A copy or cast of syntype material (compare Plastotype).</xs:documentation> </xs:annotation> </xs:enumeration> <xs:enumeration value="sec"> <xs:annotation> <xs:documentation>Secondary type -- A referred, described, measured or figured specimen in the original publication (including a neo/lectotypification publication) that is not a primary type.</xs:documentation> </xs:annotation> </xs:enumeration> <xs:enumeration value="supp"> <xs:annotation> <xs:documentation>Supplementary type -- A referred, described, measured or figured specimen in a revision of a previously described taxon.</xs:documentation> </xs:annotation> </xs:enumeration> <xs:enumeration value="syn"> <xs:annotation> <xs:documentation>Syntypes -- The series of specimens used to describe a species or infraspecific taxon when neither a single holotype by the original author, nor a lectotype in a subsequent publication has been designated. The syntypes collectively constitute the name-bearing type. [Zoo./Bot.]</xs:documentation> </xs:annotation> </xs:enumeration> <xs:enumeration value="topo"> <xs:annotation> <xs:documentation>Topotype -- One or more specimens collected at the same location as the type series (type locality), regardless of whether they are part of the type series. Topotypes are not regulated by the botanical or zoological code. Also called "locotype". [Zoo./Bot.]</xs:documentation> </xs:annotation> </xs:enumeration> <xs:enumeration value="type"> <xs:annotation> <xs:documentation>Type -- a) A specimen designated or indicated any kind of type of a species or infraspecific taxon. If possible more specific type terms (holotype, syntype, etc.) should be applied. b) the type name of a name of higher rank for taxa above the species rank. [General]</xs:documentation> </xs:annotation> </xs:enumeration> <xs:enumeration value="not"> <xs:annotation> <xs:documentation>not a type -- For specimens erroneously labelled as types an explicit negative statement may be desirable. [General]</xs:documentation> </xs:annotation> </xs:enumeration> </xs:restriction> </xs:simpleType> |
simpleType TaxonomicRankAboveSuperfamilyEnum
| namespace | http://www.tdwg.org/schemas/tcs/1.01 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| type | restriction of xs:Name | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| properties |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| used by |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| facets |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| annotation |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| source | <xs:simpleType name="TaxonomicRankAboveSuperfamilyEnum"> <xs:annotation> <xs:documentation>Subset of ranks; equivalent to BioCode "suprafamilial". This rank group includes all ranks higher than superfamily (class, phylum/division, kingdom, domain)</xs:documentation> </xs:annotation> <xs:restriction base="xs:Name"> <xs:enumeration value="infraord"> <xs:annotation> <xs:documentation>[infraord.] -- infraorder</xs:documentation> <xs:appinfo> <Specification> <PreferredAbbreviation>infraord.</PreferredAbbreviation> <AdditionalAbbreviations>-</AdditionalAbbreviations> <SuffixBacteriae>-</SuffixBacteriae> <SuffixPlantae>-</SuffixPlantae> <SuffixAlgae>-</SuffixAlgae> <SuffixFungi>-</SuffixFungi> <SuffixAnimalia>-</SuffixAnimalia> <BioCodeStatus>additional</BioCodeStatus> <BacteriaStatus>-</BacteriaStatus> <BotanyStatus>-</BotanyStatus> <ZoologyStatus>-</ZoologyStatus> </Specification> </xs:appinfo> </xs:annotation> </xs:enumeration> <xs:enumeration value="subord"> <xs:annotation> <xs:documentation>[subord.] -- suborder -- Examples: Magnolineae Catarrhini</xs:documentation> <xs:appinfo> <Specification> <PreferredAbbreviation>subord.</PreferredAbbreviation> <AdditionalAbbreviations>-</AdditionalAbbreviations> <SuffixBacteriae>-ineae</SuffixBacteriae> <SuffixPlantae>-ineae</SuffixPlantae> <SuffixAlgae>-ineae</SuffixAlgae> <SuffixFungi>-ineae</SuffixFungi> <SuffixAnimalia>-</SuffixAnimalia> <BioCodeStatus>additional</BioCodeStatus> <BacteriaStatus>covered</BacteriaStatus> <BotanyStatus>additional</BotanyStatus> <ZoologyStatus>additional</ZoologyStatus> </Specification> </xs:appinfo> </xs:annotation> </xs:enumeration> <xs:enumeration value="ord"> <xs:annotation> <xs:documentation>[ord.] -- order -- Examples: Magnoliales Primates</xs:documentation> <xs:appinfo> <Specification> <PreferredAbbreviation>ord.</PreferredAbbreviation> <AdditionalAbbreviations>-</AdditionalAbbreviations> <SuffixBacteriae>-ales</SuffixBacteriae> <SuffixPlantae>-ales</SuffixPlantae> <SuffixAlgae>-ales</SuffixAlgae> <SuffixFungi>-ales</SuffixFungi> <SuffixAnimalia>-</SuffixAnimalia> <BioCodeStatus>principal</BioCodeStatus> <BacteriaStatus>covered</BacteriaStatus> <BotanyStatus>principal</BotanyStatus> <ZoologyStatus>principal</ZoologyStatus> </Specification> </xs:appinfo> </xs:annotation> </xs:enumeration> <xs:enumeration value="superord"> <xs:annotation> <xs:documentation>[superord.] -- superorder -- Examples: Magnolianae</xs:documentation> <xs:appinfo> <Specification> <PreferredAbbreviation>superord.</PreferredAbbreviation> <AdditionalAbbreviations>-</AdditionalAbbreviations> <SuffixBacteriae>-</SuffixBacteriae> <SuffixPlantae>-anae</SuffixPlantae> <SuffixAlgae>-anae</SuffixAlgae> <SuffixFungi>-anae</SuffixFungi> <SuffixAnimalia>-</SuffixAnimalia> <BioCodeStatus>additional</BioCodeStatus> <BacteriaStatus>-</BacteriaStatus> <BotanyStatus>additional</BotanyStatus> <ZoologyStatus>additional</ZoologyStatus> </Specification> </xs:appinfo> </xs:annotation> </xs:enumeration> <xs:enumeration value="infracl"> <xs:annotation> <xs:documentation>[infracl.] -- infraclass</xs:documentation> <xs:appinfo> <Specification> <PreferredAbbreviation>infracl.</PreferredAbbreviation> <AdditionalAbbreviations>-</AdditionalAbbreviations> <SuffixBacteriae>-</SuffixBacteriae> <SuffixPlantae>-</SuffixPlantae> <SuffixAlgae>-</SuffixAlgae> <SuffixFungi>-</SuffixFungi> <SuffixAnimalia>-</SuffixAnimalia> <BioCodeStatus>additional</BioCodeStatus> <BacteriaStatus>-</BacteriaStatus> <BotanyStatus>-</BotanyStatus> <ZoologyStatus>-</ZoologyStatus> </Specification> </xs:appinfo> </xs:annotation> </xs:enumeration> <xs:enumeration value="subcl"> <xs:annotation> <xs:documentation>[subcl.] -- subclass -- Examples: Magnoliidae Eutheria</xs:documentation> <xs:appinfo> <Specification> <PreferredAbbreviation>subcl.</PreferredAbbreviation> <AdditionalAbbreviations>-</AdditionalAbbreviations> <SuffixBacteriae>-idae [proposed; Stackebrandt, E., Rainey, F.A. & Ward-Rainey, N.L.: Proposal for a new hierarchic classification system, Actinobacteria classis nov. Int. J. Syst. Bacteriol., 1997, 47, 479-491.]</SuffixBacteriae> <SuffixPlantae>-idae</SuffixPlantae> <SuffixAlgae>-phycidae</SuffixAlgae> <SuffixFungi>-mycetidae</SuffixFungi> <SuffixAnimalia>-</SuffixAnimalia> <BioCodeStatus>additional</BioCodeStatus> <BacteriaStatus>covered</BacteriaStatus> <BotanyStatus>additional</BotanyStatus> <ZoologyStatus>additional</ZoologyStatus> </Specification> </xs:appinfo> </xs:annotation> </xs:enumeration> <xs:enumeration value="cl"> <xs:annotation> <xs:documentation>[cl.] -- class -- Examples: Magnoliopsida Mammalia</xs:documentation> <xs:appinfo> <Specification> <PreferredAbbreviation>cl.</PreferredAbbreviation> <AdditionalAbbreviations>-</AdditionalAbbreviations> <SuffixBacteriae>-ia [proposed; Stackebrandt, E., Rainey, F.A. & Ward-Rainey, N.L.: Proposal for a new hierarchic classification system, Actinobacteria classis nov. Int. J. Syst. Bacteriol., 1997, 47, 479-491.]</SuffixBacteriae> <SuffixPlantae>-opsida</SuffixPlantae> <SuffixAlgae>-phyceae</SuffixAlgae> <SuffixFungi>-mycetes</SuffixFungi> <SuffixAnimalia>-</SuffixAnimalia> <BioCodeStatus>principal</BioCodeStatus> <BacteriaStatus>covered</BacteriaStatus> <BotanyStatus>principal</BotanyStatus> <ZoologyStatus>principal</ZoologyStatus> </Specification> </xs:appinfo> </xs:annotation> </xs:enumeration> <xs:enumeration value="supercl"> <xs:annotation> <xs:documentation>[supercl.] -- superclass</xs:documentation> <xs:appinfo> <Specification> <PreferredAbbreviation>supercl.</PreferredAbbreviation> <AdditionalAbbreviations>-</AdditionalAbbreviations> <SuffixBacteriae>-</SuffixBacteriae> <SuffixPlantae>-</SuffixPlantae> <SuffixAlgae>-</SuffixAlgae> <SuffixFungi>-</SuffixFungi> <SuffixAnimalia>-</SuffixAnimalia> <BioCodeStatus>additional</BioCodeStatus> <BacteriaStatus>-</BacteriaStatus> <BotanyStatus>additional</BotanyStatus> <ZoologyStatus>-</ZoologyStatus> </Specification> </xs:appinfo> </xs:annotation> </xs:enumeration> <xs:enumeration value="infraphyl_div"> <xs:annotation> <xs:documentation>[infraphyl./div.] -- infraphylum (= infradivision)</xs:documentation> <xs:appinfo> <Specification> <PreferredAbbreviation>infraphyl./div.</PreferredAbbreviation> <AdditionalAbbreviations>-</AdditionalAbbreviations> <SuffixBacteriae>-</SuffixBacteriae> <SuffixPlantae>-</SuffixPlantae> <SuffixAlgae>-</SuffixAlgae> <SuffixFungi>-</SuffixFungi> <SuffixAnimalia>-</SuffixAnimalia> <BioCodeStatus>additional</BioCodeStatus> <BacteriaStatus>-</BacteriaStatus> <BotanyStatus>-</BotanyStatus> <ZoologyStatus>-</ZoologyStatus> </Specification> </xs:appinfo> </xs:annotation> </xs:enumeration> <xs:enumeration value="subphyl_div"> <xs:annotation> <xs:documentation>[subphyl./div.] -- subphylum (= subdivision) -- Examples: Magnoliophytina Vertebrata</xs:documentation> <xs:appinfo> <Specification> <PreferredAbbreviation>subphyl./div.</PreferredAbbreviation> <AdditionalAbbreviations>-</AdditionalAbbreviations> <SuffixBacteriae>-</SuffixBacteriae> <SuffixPlantae>-phytina</SuffixPlantae> <SuffixAlgae>-phytina</SuffixAlgae> <SuffixFungi>-mycotina</SuffixFungi> <SuffixAnimalia>-</SuffixAnimalia> <BioCodeStatus>additional</BioCodeStatus> <BacteriaStatus>-</BacteriaStatus> <BotanyStatus>-</BotanyStatus> <ZoologyStatus>additional</ZoologyStatus> </Specification> </xs:appinfo> </xs:annotation> </xs:enumeration> <xs:enumeration value="phyl_div"> <xs:annotation> <xs:documentation>[phyl./div.] -- phylum (= division) -- Examples: Magnoliophyta Chordata</xs:documentation> <xs:appinfo> <Specification> <PreferredAbbreviation>phyl./div.</PreferredAbbreviation> <AdditionalAbbreviations>-</AdditionalAbbreviations> <SuffixBacteriae>-</SuffixBacteriae> <SuffixPlantae>-phyta</SuffixPlantae> <SuffixAlgae>-phyta</SuffixAlgae> <SuffixFungi>-mycota</SuffixFungi> <SuffixAnimalia>-</SuffixAnimalia> <BioCodeStatus>principal</BioCodeStatus> <BacteriaStatus>used, but all ranks above class are not covered by ICNP/ICNB</BacteriaStatus> <BotanyStatus>-</BotanyStatus> <ZoologyStatus>principal</ZoologyStatus> </Specification> </xs:appinfo> </xs:annotation> </xs:enumeration> <xs:enumeration value="superphyl_div"> <xs:annotation> <xs:documentation>[superphyl./div.] -- superphylum (= superdivision)</xs:documentation> <xs:appinfo> <Specification> <PreferredAbbreviation>superphyl./div.</PreferredAbbreviation> <AdditionalAbbreviations>-</AdditionalAbbreviations> <SuffixBacteriae>-</SuffixBacteriae> <SuffixPlantae>-</SuffixPlantae> <SuffixAlgae>-</SuffixAlgae> <SuffixFungi>-</SuffixFungi> <SuffixAnimalia>-</SuffixAnimalia> <BioCodeStatus>additional</BioCodeStatus> <BacteriaStatus>-</BacteriaStatus> <BotanyStatus>-</BotanyStatus> <ZoologyStatus>additional</ZoologyStatus> </Specification> </xs:appinfo> </xs:annotation> </xs:enumeration> <xs:enumeration value="infrareg"> <xs:annotation> <xs:documentation>[infrareg.] -- infrakingdom</xs:documentation> <xs:appinfo> <Specification> <PreferredAbbreviation>infrareg.</PreferredAbbreviation> <AdditionalAbbreviations>-</AdditionalAbbreviations> <SuffixBacteriae>-</SuffixBacteriae> <SuffixPlantae>-</SuffixPlantae> <SuffixAlgae>-</SuffixAlgae> <SuffixFungi>-</SuffixFungi> <SuffixAnimalia>-</SuffixAnimalia> <BioCodeStatus>additional</BioCodeStatus> <BacteriaStatus>-</BacteriaStatus> <BotanyStatus>-</BotanyStatus> <ZoologyStatus>-</ZoologyStatus> </Specification> </xs:appinfo> </xs:annotation> </xs:enumeration> <xs:enumeration value="subreg"> <xs:annotation> <xs:documentation>[subreg.] -- subkingdom</xs:documentation> <xs:appinfo> <Specification> <PreferredAbbreviation>subreg.</PreferredAbbreviation> <AdditionalAbbreviations>-</AdditionalAbbreviations> <SuffixBacteriae>-</SuffixBacteriae> <SuffixPlantae>-</SuffixPlantae> <SuffixAlgae>-</SuffixAlgae> <SuffixFungi>-</SuffixFungi> <SuffixAnimalia>-</SuffixAnimalia> <BioCodeStatus>additional</BioCodeStatus> <BacteriaStatus>-</BacteriaStatus> <BotanyStatus>additional</BotanyStatus> <ZoologyStatus>additional</ZoologyStatus> </Specification> </xs:appinfo> </xs:annotation> </xs:enumeration> <xs:enumeration value="reg"> <xs:annotation> <xs:documentation>[reg.] -- kingdom -- Examples: Plantae Animalia</xs:documentation> <xs:appinfo> <Specification> <PreferredAbbreviation>reg.</PreferredAbbreviation> <AdditionalAbbreviations>-</AdditionalAbbreviations> <SuffixBacteriae>-</SuffixBacteriae> <SuffixPlantae>-</SuffixPlantae> <SuffixAlgae>-</SuffixAlgae> <SuffixFungi>-</SuffixFungi> <SuffixAnimalia>-</SuffixAnimalia> <BioCodeStatus>principal</BioCodeStatus> <BacteriaStatus>used, but all ranks above class are not covered by ICNP/ICNB</BacteriaStatus> <BotanyStatus>principal</BotanyStatus> <ZoologyStatus>principal</ZoologyStatus> </Specification> </xs:appinfo> </xs:annotation> </xs:enumeration> <xs:enumeration value="superreg"> <xs:annotation> <xs:documentation>[superreg.] -- super kingdom -- Examples: Eucaryota</xs:documentation> <xs:appinfo> <Specification> <PreferredAbbreviation>superreg.</PreferredAbbreviation> <AdditionalAbbreviations>-</AdditionalAbbreviations> <SuffixBacteriae>-</SuffixBacteriae> <SuffixPlantae>-</SuffixPlantae> <SuffixAlgae>-</SuffixAlgae> <SuffixFungi>-</SuffixFungi> <SuffixAnimalia>-</SuffixAnimalia> <BioCodeStatus>additional</BioCodeStatus> <BacteriaStatus>-</BacteriaStatus> <BotanyStatus>additional</BotanyStatus> <ZoologyStatus>additional</ZoologyStatus> </Specification> </xs:appinfo> </xs:annotation> </xs:enumeration> <xs:enumeration value="dom"> <xs:annotation> <xs:documentation>[dom.] -- domain (= empire) -- Examples: Archaea (= Archaeobacteria), Bacteria (= Eubacteria), Eukarya (= Eukaryota)</xs:documentation> <xs:appinfo> <Specification> <PreferredAbbreviation>dom.</PreferredAbbreviation> <AdditionalAbbreviations>-</AdditionalAbbreviations> <SuffixBacteriae>-</SuffixBacteriae> <SuffixPlantae>-</SuffixPlantae> <SuffixAlgae>-</SuffixAlgae> <SuffixFungi>-</SuffixFungi> <SuffixAnimalia>-</SuffixAnimalia> <BioCodeStatus>secondary</BioCodeStatus> <BacteriaStatus>used, but all ranks above class are not covered by ICNP/ICNB</BacteriaStatus> <BotanyStatus>-</BotanyStatus> <ZoologyStatus>-</ZoologyStatus> </Specification> </xs:appinfo> </xs:annotation> </xs:enumeration> <xs:enumeration value="taxsupragen"> <xs:annotation> <xs:documentation>[tax. supragen.] -- suprageneric tax. of undefined rank -- This value indicates that the rank of a name is unknown. Compare "incertae sedis" which is commonly used as a replacement for a taxon to group all taxa whose position in the classification or phylogenetic tree is uncertain.</xs:documentation> <xs:appinfo> <Specification> <PreferredAbbreviation>tax. supragen.</PreferredAbbreviation> <AdditionalAbbreviations>-</AdditionalAbbreviations> <SuffixBacteriae>-</SuffixBacteriae> <SuffixPlantae>-</SuffixPlantae> <SuffixAlgae>-</SuffixAlgae> <SuffixFungi>-</SuffixFungi> <SuffixAnimalia>-</SuffixAnimalia> <BioCodeStatus>-</BioCodeStatus> <BacteriaStatus>-</BacteriaStatus> <BotanyStatus>-</BotanyStatus> <ZoologyStatus>-</ZoologyStatus> </Specification> </xs:appinfo> </xs:annotation> </xs:enumeration> </xs:restriction> </xs:simpleType> |
simpleType TaxonomicRankBelowSubspeciesEnum
| namespace | http://www.tdwg.org/schemas/tcs/1.01 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| type | restriction of xs:Name | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| properties |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| used by |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| facets |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| annotation |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| source | <xs:simpleType name="TaxonomicRankBelowSubspeciesEnum"> <xs:annotation> <xs:documentation>Subset of ranks; equivalent to BioCode "infra-subspecfic", i.e. below the species group</xs:documentation> </xs:annotation> <xs:restriction base="xs:Name"> <xs:enumeration value="cand"> <xs:annotation> <xs:documentation>[cand.] -- candidate -- Candidatus' rank is proposed in bacteriology for putative taxa, which could not yet be studied sufficiently to warrant the creation of a name with a known rank. (Murray, R.G.E. & Schleifer, K.H.: Taxonomic notes: a proposal for recording the properties of putative taxa of procaryotes. Int. J. Syst. Bacteriol., 1994, 44, 174-176).</xs:documentation> <xs:appinfo> <Specification> <PreferredAbbreviation>cand.</PreferredAbbreviation> <AdditionalAbbreviations>-</AdditionalAbbreviations> <SuffixBacteriae>-</SuffixBacteriae> <SuffixPlantae>-</SuffixPlantae> <SuffixAlgae>-</SuffixAlgae> <SuffixFungi>-</SuffixFungi> <SuffixAnimalia>-</SuffixAnimalia> <BioCodeStatus>-</BioCodeStatus> <BacteriaStatus>Proposed</BacteriaStatus> <BotanyStatus>-</BotanyStatus> <ZoologyStatus>-</ZoologyStatus> </Specification> </xs:appinfo> </xs:annotation> </xs:enumeration> <xs:enumeration value="taxinfrasp"> <xs:annotation> <xs:documentation>[tax. infrasp.] -- infraspecific tax. of undefined rank -- Undefined ranks (using either no rank identifier in botany, or using greek letters or symbols like stars, crosses) occur in very old publications. Most frequently these are to be interpreted as varieties, but occasionally they are forms or subspecies (see Stearn, W.T. 1957: Species plantarum (Facsimile); Introduction. 1. London, p. 90-95, 160-161, 163). The interpretation of these cases requires taxonomic knowledge that may not be available at the time when data are parsed. Such lack of knowledge can be expressed using this rank identifier.</xs:documentation> <xs:appinfo> <Specification> <PreferredAbbreviation>tax. infrasp.</PreferredAbbreviation> <AdditionalAbbreviations>-</AdditionalAbbreviations> <SuffixBacteriae>-</SuffixBacteriae> <SuffixPlantae>-</SuffixPlantae> <SuffixAlgae>-</SuffixAlgae> <SuffixFungi>-</SuffixFungi> <SuffixAnimalia>-</SuffixAnimalia> <BioCodeStatus>-</BioCodeStatus> <BacteriaStatus>-</BacteriaStatus> <BotanyStatus>-</BotanyStatus> <ZoologyStatus>-</ZoologyStatus> </Specification> </xs:appinfo> </xs:annotation> </xs:enumeration> <xs:enumeration value="fsp"> <xs:annotation> <xs:documentation>[f. sp.] -- special form -- The ICBN does not formally cover formae specialis (art. 4, note 3). However, because of the economic importance of pathogenic f. sp., and since it is common practice to handle them as if the code would apply (i. e. priority usually observed, name quoted with author), they are included here.</xs:documentation> <xs:appinfo> <Specification> <PreferredAbbreviation>f. sp.</PreferredAbbreviation> <AdditionalAbbreviations>forma sp.; fsp.; fm. sp.; f. spec.; fm. spec.; forma spec.</AdditionalAbbreviations> <SuffixBacteriae>-</SuffixBacteriae> <SuffixPlantae>-</SuffixPlantae> <SuffixAlgae>-</SuffixAlgae> <SuffixFungi>-</SuffixFungi> <SuffixAnimalia>-</SuffixAnimalia> <BioCodeStatus>-</BioCodeStatus> <BacteriaStatus>used, but all ranks below subsp. are not covered by ICNP/ICNB, see Rules 5d and 14a. ##Check whether this rank is used indeed.</BacteriaStatus> <BotanyStatus>none</BotanyStatus> <ZoologyStatus>-</ZoologyStatus> </Specification> </xs:appinfo> </xs:annotation> </xs:enumeration> <xs:enumeration value="subsubfm"> <xs:annotation> <xs:documentation>[subsubfm.] -- subsubform</xs:documentation> <xs:appinfo> <Specification> <PreferredAbbreviation>subsubfm.</PreferredAbbreviation> <AdditionalAbbreviations>subsubf.</AdditionalAbbreviations> <SuffixBacteriae>-</SuffixBacteriae> <SuffixPlantae>-</SuffixPlantae> <SuffixAlgae>-</SuffixAlgae> <SuffixFungi>-</SuffixFungi> <SuffixAnimalia>-</SuffixAnimalia> <BioCodeStatus>-</BioCodeStatus> <BacteriaStatus>-</BacteriaStatus> <BotanyStatus>additional(?)</BotanyStatus> <ZoologyStatus>-</ZoologyStatus> </Specification> </xs:appinfo> </xs:annotation> </xs:enumeration> <xs:enumeration value="subfm"> <xs:annotation> <xs:documentation>[subfm.] -- subform</xs:documentation> <xs:appinfo> <Specification> <PreferredAbbreviation>subfm.</PreferredAbbreviation> <AdditionalAbbreviations>subf.</AdditionalAbbreviations> <SuffixBacteriae>-</SuffixBacteriae> <SuffixPlantae>-</SuffixPlantae> <SuffixAlgae>-</SuffixAlgae> <SuffixFungi>-</SuffixFungi> <SuffixAnimalia>-</SuffixAnimalia> <BioCodeStatus>additional</BioCodeStatus> <BacteriaStatus>-</BacteriaStatus> <BotanyStatus>additional</BotanyStatus> <ZoologyStatus>-</ZoologyStatus> </Specification> </xs:appinfo> </xs:annotation> </xs:enumeration> <xs:enumeration value="fm"> <xs:annotation> <xs:documentation>[fm.] -- form -- Form, race, variety are not subject to regulation in zoology; see ICZN Article 1.3.4 </xs:documentation> <xs:appinfo> <Specification> <PreferredAbbreviation>fm.</PreferredAbbreviation> <AdditionalAbbreviations>f.</AdditionalAbbreviations> <SuffixBacteriae>-</SuffixBacteriae> <SuffixPlantae>-</SuffixPlantae> <SuffixAlgae>-</SuffixAlgae> <SuffixFungi>-</SuffixFungi> <SuffixAnimalia>-</SuffixAnimalia> <BioCodeStatus>secondary</BioCodeStatus> <BacteriaStatus>-</BacteriaStatus> <BotanyStatus>secondary</BotanyStatus> <ZoologyStatus>-</ZoologyStatus> </Specification> </xs:appinfo> </xs:annotation> </xs:enumeration> <xs:enumeration value="subsubvar"> <xs:annotation> <xs:documentation>[subsubvar.] -- sub-sub-variety</xs:documentation> <xs:appinfo> <Specification> <PreferredAbbreviation>subsubvar.</PreferredAbbreviation> <AdditionalAbbreviations>subsubv.</AdditionalAbbreviations> <SuffixBacteriae>-</SuffixBacteriae> <SuffixPlantae>-</SuffixPlantae> <SuffixAlgae>-</SuffixAlgae> <SuffixFungi>-</SuffixFungi> <SuffixAnimalia>-</SuffixAnimalia> <BioCodeStatus>additional</BioCodeStatus> <BacteriaStatus>-</BacteriaStatus> <BotanyStatus>additional</BotanyStatus> <ZoologyStatus>-</ZoologyStatus> </Specification> </xs:appinfo> </xs:annotation> </xs:enumeration> <xs:enumeration value="subvar"> <xs:annotation> <xs:documentation>[subvar.] -- sub-variety</xs:documentation> <xs:appinfo> <Specification> <PreferredAbbreviation>subvar.</PreferredAbbreviation> <AdditionalAbbreviations>subv.</AdditionalAbbreviations> <SuffixBacteriae>-</SuffixBacteriae> <SuffixPlantae>-</SuffixPlantae> <SuffixAlgae>-</SuffixAlgae> <SuffixFungi>-</SuffixFungi> <SuffixAnimalia>-</SuffixAnimalia> <BioCodeStatus>additional</BioCodeStatus> <BacteriaStatus>-</BacteriaStatus> <BotanyStatus>additional</BotanyStatus> <ZoologyStatus>-</ZoologyStatus> </Specification> </xs:appinfo> </xs:annotation> </xs:enumeration> <xs:enumeration value="var"> <xs:annotation> <xs:documentation>[var.] -- variety -- Form, race, variety are not subject to regulation in zoology; see ICZN Article 1.3.4 Examples: Pinus nigra var. caramanica (= "P. nigra subsp. nigra var. caramanica"; Taxus baccata var. variegata</xs:documentation> <xs:appinfo> <Specification> <PreferredAbbreviation>var.</PreferredAbbreviation> <AdditionalAbbreviations>v.</AdditionalAbbreviations> <SuffixBacteriae>-</SuffixBacteriae> <SuffixPlantae>-</SuffixPlantae> <SuffixAlgae>-</SuffixAlgae> <SuffixFungi>-</SuffixFungi> <SuffixAnimalia>-</SuffixAnimalia> <BioCodeStatus>secondary</BioCodeStatus> <BacteriaStatus>used, but all ranks below subsp. are not covered by ICNP/ICNB, see Rules 5d and 14a. ##Check whether this rank is used indeed.</BacteriaStatus> <BotanyStatus>secondary</BotanyStatus> <ZoologyStatus>-</ZoologyStatus> </Specification> </xs:appinfo> </xs:annotation> </xs:enumeration> <xs:enumeration value="pv"> <xs:annotation> <xs:documentation>[pathovar.] -- patho-variety</xs:documentation> <xs:appinfo> <Specification> <PreferredAbbreviation>pathovar.</PreferredAbbreviation> <AdditionalAbbreviations>pv.</AdditionalAbbreviations> <SuffixBacteriae>-</SuffixBacteriae> <SuffixPlantae>-</SuffixPlantae> <SuffixAlgae>-</SuffixAlgae> <SuffixFungi>-</SuffixFungi> <SuffixAnimalia>-</SuffixAnimalia> <BioCodeStatus>-</BioCodeStatus> <BacteriaStatus>used, but all ranks below subsp. are not covered by ICNP/ICNB, see Rules 5d and 14a. ##Check whether this rank is used indeed.</BacteriaStatus> <BotanyStatus>-</BotanyStatus> <ZoologyStatus>-</ZoologyStatus> </Specification> </xs:appinfo> </xs:annotation> </xs:enumeration> <xs:enumeration value="bv"> <xs:annotation> <xs:documentation>[biovar.] -- bio-variety</xs:documentation> <xs:appinfo> <Specification> <PreferredAbbreviation>biovar.</PreferredAbbreviation> <AdditionalAbbreviations>bv.</AdditionalAbbreviations> <SuffixBacteriae>-</SuffixBacteriae> <SuffixPlantae>-</SuffixPlantae> <SuffixAlgae>-</SuffixAlgae> <SuffixFungi>-</SuffixFungi> <SuffixAnimalia>-</SuffixAnimalia> <BioCodeStatus>-</BioCodeStatus> <BacteriaStatus>used, but all ranks below subsp. are not covered by ICNP/ICNB, see Rules 5d and 14a. ##Check whether this rank is used indeed.</BacteriaStatus> <BotanyStatus>-</BotanyStatus> <ZoologyStatus>-</ZoologyStatus> </Specification> </xs:appinfo> </xs:annotation> </xs:enumeration> <xs:enumeration value="infrasp"> <xs:annotation> <xs:documentation>[infrasp.] -- infraspecies</xs:documentation> <xs:appinfo> <Specification> <PreferredAbbreviation>infrasp.</PreferredAbbreviation> <AdditionalAbbreviations>infrasp.; infraspec.</AdditionalAbbreviations> <SuffixBacteriae>-</SuffixBacteriae> <SuffixPlantae>-</SuffixPlantae> <SuffixAlgae>-</SuffixAlgae> <SuffixFungi>-</SuffixFungi> <SuffixAnimalia>-</SuffixAnimalia> <BioCodeStatus>additional</BioCodeStatus> <BacteriaStatus>-</BacteriaStatus> <BotanyStatus>-</BotanyStatus> <ZoologyStatus>-</ZoologyStatus> </Specification> </xs:appinfo> </xs:annotation> </xs:enumeration> </xs:restriction> </xs:simpleType> |
simpleType TaxonomicRankCultivatedPlants
| namespace | http://www.tdwg.org/schemas/tcs/1.01 | |||||||||||||||||||||||||||||||||||||||||||||
| type | restriction of xs:Name | |||||||||||||||||||||||||||||||||||||||||||||
| properties |
|
|||||||||||||||||||||||||||||||||||||||||||||
| used by |
|
|||||||||||||||||||||||||||||||||||||||||||||
| facets |
|
|||||||||||||||||||||||||||||||||||||||||||||
| annotation |
|
|||||||||||||||||||||||||||||||||||||||||||||
| source | <xs:simpleType name="TaxonomicRankCultivatedPlants"> <xs:annotation> <xs:documentation>Subset of ranks used spefically for cultivated plants</xs:documentation> </xs:annotation> <xs:restriction base="xs:Name"> <xs:enumeration value="cv"> <xs:annotation> <xs:documentation>[cult.] -- cultivar -- The epithet is usually output in single quotes and may contain multiple words, see ICBN §28. Examples: Taxus baccata 'Variegata', Juniperus ×pfitzeriana 'Wilhelm Pfitzer'; Magnolia 'Elizabeth' (= a hybrid, no species epithet). </xs:documentation> <xs:appinfo> <Specification> <PreferredAbbreviation>cult.</PreferredAbbreviation> <AdditionalAbbreviations>-</AdditionalAbbreviations> <SuffixBacteriae>-</SuffixBacteriae> <SuffixPlantae>-</SuffixPlantae> <SuffixAlgae>-</SuffixAlgae> <SuffixFungi>-</SuffixFungi> <SuffixAnimalia>-</SuffixAnimalia> <BioCodeStatus>-</BioCodeStatus> <BacteriaStatus>-</BacteriaStatus> <BotanyStatus>Reference to 'Internat. code of nomenclature for cultivated plants'</BotanyStatus> <ZoologyStatus>-</ZoologyStatus> </Specification> </xs:appinfo> </xs:annotation> </xs:enumeration> <xs:enumeration value="convar"> <xs:annotation> <xs:documentation>[convar.] -- convar -- Used in cultivated plants (ICNCP), but deprecated, see 'Some notes on problems of taxonomy and nomenclature of cultivated plants' by J. Ochsmann, http://www.genres.de/IGRREIHE/IGRREIHE/DDD/22-08.pdf</xs:documentation> <xs:appinfo> <Specification> <PreferredAbbreviation>convar.</PreferredAbbreviation> <AdditionalAbbreviations>cv.</AdditionalAbbreviations> <SuffixBacteriae>-</SuffixBacteriae> <SuffixPlantae>-</SuffixPlantae> <SuffixAlgae>-</SuffixAlgae> <SuffixFungi>-</SuffixFungi> <SuffixAnimalia>-</SuffixAnimalia> <BioCodeStatus>-</BioCodeStatus> <BacteriaStatus>-</BacteriaStatus> <BotanyStatus>-</BotanyStatus> <ZoologyStatus>-</ZoologyStatus> </Specification> </xs:appinfo> </xs:annotation> </xs:enumeration> <xs:enumeration value="grex"> <xs:annotation> <xs:documentation> [grex] -- a rank equivalent to cultivar group used principly for artificial hybrid swams in Orchidaceae. </xs:documentation> <xs:appinfo> <Specification> <PreferredAbbreviation>grex</PreferredAbbreviation> <AdditionalAbbreviations>grex</AdditionalAbbreviations> <SuffixBacteriae>-</SuffixBacteriae> <SuffixPlantae>-</SuffixPlantae> <SuffixAlgae>-</SuffixAlgae> <SuffixFungi>-</SuffixFungi> <SuffixAnimalia>-</SuffixAnimalia> <BioCodeStatus>-</BioCodeStatus> <BacteriaStatus>-</BacteriaStatus> <BotanyStatus>Reference to ICNCP</BotanyStatus> <ZoologyStatus>-</ZoologyStatus> </Specification> </xs:appinfo> </xs:annotation> </xs:enumeration> <xs:enumeration value="cvgroup"> <xs:annotation> <xs:documentation>[cultivar. group] -- cultivar-group</xs:documentation> <xs:appinfo> <Specification> <PreferredAbbreviation>cultivar. group</PreferredAbbreviation> <AdditionalAbbreviations>-</AdditionalAbbreviations> <SuffixBacteriae>-</SuffixBacteriae> <SuffixPlantae>-</SuffixPlantae> <SuffixAlgae>-</SuffixAlgae> <SuffixFungi>-</SuffixFungi> <SuffixAnimalia>-</SuffixAnimalia> <BioCodeStatus>-</BioCodeStatus> <BacteriaStatus>-</BacteriaStatus> <BotanyStatus>-</BotanyStatus> <ZoologyStatus>-</ZoologyStatus> </Specification> </xs:appinfo> </xs:annotation> </xs:enumeration> <xs:enumeration value="graftchimaera"> <xs:annotation> <xs:documentation>[graft-chimaera] -- graft-chimaera</xs:documentation> <xs:appinfo> <Specification> <PreferredAbbreviation>graft-chimaera</PreferredAbbreviation> <AdditionalAbbreviations>-</AdditionalAbbreviations> <SuffixBacteriae>-</SuffixBacteriae> <SuffixPlantae>-</SuffixPlantae> <SuffixAlgae>-</SuffixAlgae> <SuffixFungi>-</SuffixFungi> <SuffixAnimalia>-</SuffixAnimalia> <BioCodeStatus>-</BioCodeStatus> <BacteriaStatus>-</BacteriaStatus> <BotanyStatus>-</BotanyStatus> <ZoologyStatus>-</ZoologyStatus> </Specification> </xs:appinfo> </xs:annotation> </xs:enumeration> <xs:enumeration value="denomclass"> <xs:annotation> <xs:documentation>Denomination classes are constructs under ICNCP for defining the scope within which a cultivar or Group epithet must be unique.</xs:documentation> <xs:appinfo> <Specification> <PreferredAbbreviation>denomination-class</PreferredAbbreviation> <AdditionalAbbreviations>-</AdditionalAbbreviations> <SuffixBacteriae>-</SuffixBacteriae> <SuffixPlantae>-</SuffixPlantae> <SuffixAlgae>-</SuffixAlgae> <SuffixFungi>-</SuffixFungi> <SuffixAnimalia>-</SuffixAnimalia> <BioCodeStatus>-</BioCodeStatus> <BacteriaStatus>-</BacteriaStatus> <BotanyStatus>-</BotanyStatus> <ZoologyStatus>-</ZoologyStatus> </Specification> </xs:appinfo> </xs:annotation> </xs:enumeration> </xs:restriction> </xs:simpleType> |
simpleType TaxonomicRankEnum
| namespace | http://www.tdwg.org/schemas/tcs/1.01 | ||
| type | union of (TaxonomicRankBelowSubspeciesEnum, TaxonomicRankSpeciesGroupEnum, TaxonomicRankGenusSubdivisionEnum, TaxonomicRankGenusGroupEnum, TaxonomicRankFamilySubdivisionEnum, TaxonomicRankFamilyGroupEnum, TaxonomicRankAboveSuperfamilyEnum, TaxonomicRankCultivatedPlants) | ||
| used by |
|
||
| annotation |
|
||
| source | <xs:simpleType name="TaxonomicRankEnum"> <xs:annotation> <xs:documentation> <p>Enumerated codes to express the rank of a taxon (scientific organism name) in a taxonomic hierarchy. The list is intended to be interoperable between name providers for bacteria, viruses, fungi, plants, and animals. It is not assumed that in each taxonomic group all ranks have to be used. Individual applications may select appropriate subsets (which may be based on information given inside the enumerated values, see Specifications/BioCode-, Botany-, Zoology-, and BacteriaStatus). The enumeration attempts to strike a balance between listing all possible rank terms, and remaining comprehensible. For example, the "infra-" ranks specifically mentioned in BioCode have been included (although very rarely used), but the additional intermediate zoological ranks (micro, nano, pico, etc.) are not included. Whether the selection of infraspecific ranks (some informal ranks, esp. from bacteriology, may be missing!) probably needs some discussion. However, it is believed that this list may help to start developing data sets that can easily be integrated across the barriers of Language and taxonomic traditions.</p> <p>Not included in the list are the botanical "notho-" ranks, which are used to designate hybrids (nothospecies, nothogenus). It is assumed they can be generated from separate information that the taxon is a hybrid. ICBN §4.4 states: "The subordinate ranks of nothotaxa are the same as the subordinate ranks of non-hybrid taxa, except that nothogenus is the highest rank permitted".</p> <p>The following publications have been consulted to determine the number of type terms that should be included and to prepare the semantic definitions:</p> <ul> <li>The Berlin Taxonomic Information Model, MoReTax view (Berendsohn & al., http://www.bgbm.org/scripts/ASP/BGBMModel/Catalogues.asp?Cat=MT</li> <li>DiversityTaxonomy model version 0.7 (G. Hagedorn & T. Grafenhan 2002, http://160.45.63.11/Workbench/Taxonomy/Model/InformationModels.html)</li> <li>ABCD version 1.44, types HigherTaxonRankType and RankAbbreviationType, by W. Berendsohn, reviewed by D. Hobern</li> <li>TaxCat2 - Database of Botanical Taxonomic Categories by Jorg Ochsmann, IPK Gatersleben; http://mansfeld.ipk-gatersleben.de/TaxCat2/default.htm</li> </ul> <p>A separate enumeration and several ranks have been added to the original list to better accommodate names from ICNCP. (RDH). </p> <p>Many thanks for review and help go to Dr. Walter Gams.</p> <p>Note: the list of all ranks is implemented as a union of all following rank subsets. Note that although BioCode has been used to define the partition into subsets, the ranks are not limited to BioCode but should be an interoperable superset of ranks used in Virology, Bacteriology, Botany and Zoology.</p> </xs:documentation> </xs:annotation> <xs:union memberTypes="TaxonomicRankBelowSubspeciesEnum TaxonomicRankSpeciesGroupEnum TaxonomicRankGenusSubdivisionEnum TaxonomicRankGenusGroupEnum TaxonomicRankFamilySubdivisionEnum TaxonomicRankFamilyGroupEnum TaxonomicRankAboveSuperfamilyEnum TaxonomicRankCultivatedPlants"/> </xs:simpleType> |
simpleType TaxonomicRankFamilyGroupEnum
| namespace | http://www.tdwg.org/schemas/tcs/1.01 | |||||||||||||||||||||||||||||||
| type | restriction of xs:Name | |||||||||||||||||||||||||||||||
| properties |
|
|||||||||||||||||||||||||||||||
| used by |
|
|||||||||||||||||||||||||||||||
| facets |
|
|||||||||||||||||||||||||||||||
| annotation |
|
|||||||||||||||||||||||||||||||
| source | <xs:simpleType name="TaxonomicRankFamilyGroupEnum"> <xs:annotation> <xs:documentation>Subset of ranks; equivalent to BioCode "family group", i.e. infrafamily to superfamily</xs:documentation> </xs:annotation> <xs:restriction base="xs:Name"> <xs:enumeration value="infrafam"> <xs:annotation> <xs:documentation>[infrafam.] -- infrafamily</xs:documentation> <xs:appinfo> <Specification> <PreferredAbbreviation>infrafam.</PreferredAbbreviation> <AdditionalAbbreviations>-</AdditionalAbbreviations> <SuffixBacteriae>-</SuffixBacteriae> <SuffixPlantae>-</SuffixPlantae> <SuffixAlgae>-</SuffixAlgae> <SuffixFungi>-</SuffixFungi> <SuffixAnimalia>-</SuffixAnimalia> <BioCodeStatus>additional</BioCodeStatus> <BacteriaStatus>-</BacteriaStatus> <BotanyStatus>-</BotanyStatus> <ZoologyStatus>-</ZoologyStatus> </Specification> </xs:appinfo> </xs:annotation> </xs:enumeration> <xs:enumeration value="subfam"> <xs:annotation> <xs:documentation>[subfam.] -- subfamily -- Examples: Magnolioideae</xs:documentation> <xs:appinfo> <Specification> <PreferredAbbreviation>subfam.</PreferredAbbreviation> <AdditionalAbbreviations>-</AdditionalAbbreviations> <SuffixBacteriae>-oideae</SuffixBacteriae> <SuffixPlantae>-oideae</SuffixPlantae> <SuffixAlgae>-oideae</SuffixAlgae> <SuffixFungi>-oideae</SuffixFungi> <SuffixAnimalia>-inae</SuffixAnimalia> <BioCodeStatus>secondary</BioCodeStatus> <BacteriaStatus>covered</BacteriaStatus> <BotanyStatus>additional</BotanyStatus> <ZoologyStatus>additional</ZoologyStatus> </Specification> </xs:appinfo> </xs:annotation> </xs:enumeration> <xs:enumeration value="fam"> <xs:annotation> <xs:documentation>[fam.] -- family -- Examples: Magnoliaceae Hominidae</xs:documentation> <xs:appinfo> <Specification> <PreferredAbbreviation>fam.</PreferredAbbreviation> <AdditionalAbbreviations>-</AdditionalAbbreviations> <SuffixBacteriae>-aceae</SuffixBacteriae> <SuffixPlantae>-aceae</SuffixPlantae> <SuffixAlgae>-aceae</SuffixAlgae> <SuffixFungi>-aceae</SuffixFungi> <SuffixAnimalia>-idae</SuffixAnimalia> <BioCodeStatus>principal</BioCodeStatus> <BacteriaStatus>covered</BacteriaStatus> <BotanyStatus>principal</BotanyStatus> <ZoologyStatus>principal</ZoologyStatus> </Specification> </xs:appinfo> </xs:annotation> </xs:enumeration> <xs:enumeration value="superfam"> <xs:annotation> <xs:documentation>[superfam.] -- superfamily -- Examples: Magnoliacea</xs:documentation> <xs:appinfo> <Specification> <PreferredAbbreviation>superfam.</PreferredAbbreviation> <AdditionalAbbreviations>-</AdditionalAbbreviations> <SuffixBacteriae>-</SuffixBacteriae> <SuffixPlantae>-acea</SuffixPlantae> <SuffixAlgae>-acea</SuffixAlgae> <SuffixFungi>-acea</SuffixFungi> <SuffixAnimalia>-oidea; -acea</SuffixAnimalia> <BioCodeStatus>secondary</BioCodeStatus> <BacteriaStatus>-</BacteriaStatus> <BotanyStatus>additional</BotanyStatus> <ZoologyStatus>-</ZoologyStatus> </Specification> </xs:appinfo> </xs:annotation> </xs:enumeration> </xs:restriction> </xs:simpleType> |
simpleType TaxonomicRankFamilySubdivisionEnum
| namespace | http://www.tdwg.org/schemas/tcs/1.01 | |||||||||||||||||||||||||||||||
| type | restriction of xs:Name | |||||||||||||||||||||||||||||||
| properties |
|
|||||||||||||||||||||||||||||||
| used by |
|
|||||||||||||||||||||||||||||||
| facets |
|
|||||||||||||||||||||||||||||||
| annotation |
|
|||||||||||||||||||||||||||||||
| source | <xs:simpleType name="TaxonomicRankFamilySubdivisionEnum"> <xs:annotation> <xs:documentation>Subset of ranks; equivalent to BioCode "subdivision of a family", i.e. ranks between genus group and family group</xs:documentation> </xs:annotation> <xs:restriction base="xs:Name"> <xs:enumeration value="infratrib"> <xs:annotation> <xs:documentation>[infratrib.] -- infratribe</xs:documentation> <xs:appinfo> <Specification> <PreferredAbbreviation>infratrib.</PreferredAbbreviation> <AdditionalAbbreviations>-</AdditionalAbbreviations> <SuffixBacteriae>-</SuffixBacteriae> <SuffixPlantae>-</SuffixPlantae> <SuffixAlgae>-</SuffixAlgae> <SuffixFungi>-</SuffixFungi> <SuffixAnimalia>-</SuffixAnimalia> <BioCodeStatus>additional</BioCodeStatus> <BacteriaStatus>-</BacteriaStatus> <BotanyStatus>-</BotanyStatus> <ZoologyStatus>-</ZoologyStatus> </Specification> </xs:appinfo> </xs:annotation> </xs:enumeration> <xs:enumeration value="subtrib"> <xs:annotation> <xs:documentation>[subtrib.] -- subtribe</xs:documentation> <xs:appinfo> <Specification> <PreferredAbbreviation>subtrib.</PreferredAbbreviation> <AdditionalAbbreviations>-</AdditionalAbbreviations> <SuffixBacteriae>-inae</SuffixBacteriae> <SuffixPlantae>-inae</SuffixPlantae> <SuffixAlgae>-inae</SuffixAlgae> <SuffixFungi>-inae</SuffixFungi> <SuffixAnimalia>-ina</SuffixAnimalia> <BioCodeStatus>additional</BioCodeStatus> <BacteriaStatus>covered (but probably not in current use)</BacteriaStatus> <BotanyStatus>additional</BotanyStatus> <ZoologyStatus>-</ZoologyStatus> </Specification> </xs:appinfo> </xs:annotation> </xs:enumeration> <xs:enumeration value="trib"> <xs:annotation> <xs:documentation>[trib.] -- tribe</xs:documentation> <xs:appinfo> <Specification> <PreferredAbbreviation>trib.</PreferredAbbreviation> <AdditionalAbbreviations>-</AdditionalAbbreviations> <SuffixBacteriae>-eae</SuffixBacteriae> <SuffixPlantae>-eae</SuffixPlantae> <SuffixAlgae>-eae</SuffixAlgae> <SuffixFungi>-eae</SuffixFungi> <SuffixAnimalia>-ini</SuffixAnimalia> <BioCodeStatus>secondary</BioCodeStatus> <BacteriaStatus>covered (but probably not in current use)</BacteriaStatus> <BotanyStatus>secondary</BotanyStatus> <ZoologyStatus>-</ZoologyStatus> </Specification> </xs:appinfo> </xs:annotation> </xs:enumeration> <xs:enumeration value="supertrib"> <xs:annotation> <xs:documentation>[supertrib.] -- supertribe</xs:documentation> <xs:appinfo> <Specification> <PreferredAbbreviation>supertrib.</PreferredAbbreviation> <AdditionalAbbreviations>-</AdditionalAbbreviations> <SuffixBacteriae>-</SuffixBacteriae> <SuffixPlantae>-</SuffixPlantae> <SuffixAlgae>-</SuffixAlgae> <SuffixFungi>-</SuffixFungi> <SuffixAnimalia>-</SuffixAnimalia> <BioCodeStatus>additional</BioCodeStatus> <BacteriaStatus>-</BacteriaStatus> <BotanyStatus>additional</BotanyStatus> <ZoologyStatus>-</ZoologyStatus> </Specification> </xs:appinfo> </xs:annotation> </xs:enumeration> </xs:restriction> </xs:simpleType> |
simpleType TaxonomicRankGenusGroupEnum
| namespace | http://www.tdwg.org/schemas/tcs/1.01 | ||||||||||||||||||||||||
| type | restriction of xs:Name | ||||||||||||||||||||||||
| properties |
|
||||||||||||||||||||||||
| used by |
|
||||||||||||||||||||||||
| facets |
|
||||||||||||||||||||||||
| annotation |
|
||||||||||||||||||||||||
| source | <xs:simpleType name="TaxonomicRankGenusGroupEnum"> <xs:annotation> <xs:documentation>Subset of ranks; equivalent to BioCode "genus group", i.e. infragenus to genus</xs:documentation> </xs:annotation> <xs:restriction base="xs:Name"> <xs:enumeration value="infragen"> <xs:annotation> <xs:documentation>[infragen.] -- infragenus</xs:documentation> <xs:appinfo> <Specification> <PreferredAbbreviation>infragen.</PreferredAbbreviation> <AdditionalAbbreviations>*</AdditionalAbbreviations> <SuffixBacteriae>-</SuffixBacteriae> <SuffixPlantae>-</SuffixPlantae> <SuffixAlgae>-</SuffixAlgae> <SuffixFungi>-</SuffixFungi> <SuffixAnimalia>-</SuffixAnimalia> <BioCodeStatus>additional</BioCodeStatus> <BacteriaStatus>-</BacteriaStatus> <BotanyStatus>-</BotanyStatus> <ZoologyStatus>-</ZoologyStatus> </Specification> </xs:appinfo> </xs:annotation> </xs:enumeration> <xs:enumeration value="subgen"> <xs:annotation> <xs:documentation>[subgen.] -- subgenus</xs:documentation> <xs:appinfo> <Specification> <PreferredAbbreviation>subgen.</PreferredAbbreviation> <AdditionalAbbreviations>*</AdditionalAbbreviations> <SuffixBacteriae>-</SuffixBacteriae> <SuffixPlantae>-</SuffixPlantae> <SuffixAlgae>-</SuffixAlgae> <SuffixFungi>-</SuffixFungi> <SuffixAnimalia>-</SuffixAnimalia> <BioCodeStatus>secondary</BioCodeStatus> <BacteriaStatus>covered</BacteriaStatus> <BotanyStatus>additional</BotanyStatus> <ZoologyStatus>additional</ZoologyStatus> </Specification> </xs:appinfo> </xs:annotation> </xs:enumeration> <xs:enumeration value="gen"> <xs:annotation> <xs:documentation>[gen.] -- genus -- Examples: Magnolia Homo</xs:documentation> <xs:appinfo> <Specification> <PreferredAbbreviation>gen.</PreferredAbbreviation> <AdditionalAbbreviations>-</AdditionalAbbreviations> <SuffixBacteriae>-</SuffixBacteriae> <SuffixPlantae>-</SuffixPlantae> <SuffixAlgae>-</SuffixAlgae> <SuffixFungi>-</SuffixFungi> <SuffixAnimalia>-</SuffixAnimalia> <BioCodeStatus>primary</BioCodeStatus> <BacteriaStatus>covered</BacteriaStatus> <BotanyStatus>principal</BotanyStatus> <ZoologyStatus>principal</ZoologyStatus> </Specification> </xs:appinfo> </xs:annotation> </xs:enumeration> </xs:restriction> </xs:simpleType> |
simpleType TaxonomicRankGenusSubdivisionEnum
| namespace | http://www.tdwg.org/schemas/tcs/1.01 | |||||||||||||||||||||||||||||||||||||||||||||
| type | restriction of xs:Name | |||||||||||||||||||||||||||||||||||||||||||||
| properties |
|
|||||||||||||||||||||||||||||||||||||||||||||
| used by |
|
|||||||||||||||||||||||||||||||||||||||||||||
| facets |
|
|||||||||||||||||||||||||||||||||||||||||||||
| annotation |
|
|||||||||||||||||||||||||||||||||||||||||||||
| source | <xs:simpleType name="TaxonomicRankGenusSubdivisionEnum"> <xs:annotation> <xs:documentation>Subset of ranks; equivalent to BioCode ""subdivision of a genus" ", i.e. all ranks between genus and species group (i.e. not including subgenus and species)</xs:documentation> </xs:annotation> <xs:restriction base="xs:Name"> <xs:enumeration value="aggr"> <xs:annotation> <xs:documentation>[aggr.] -- species aggregate (= species group, species complex) -- A loosely defined group of species. Zoology: "Aggregate - a group of species, other than a subgenus, within a genus. An aggregate may be denoted by a group name interpolated in parentheses." -- The Berlin/MoreTax model notes: "[these] aren't taxonomic ranks but cirumscriptions because on the one hand they are necessary for the concatenation of the fullname and on the other hand they are necessary for distinguishing the aggregate or species group from the microspecies." Compare subspecific aggregate for a group of subspecies within a species!</xs:documentation> <xs:appinfo> <Specification> <PreferredAbbreviation>aggr.</PreferredAbbreviation> <AdditionalAbbreviations/> <SuffixBacteriae>-</SuffixBacteriae> <SuffixPlantae>-</SuffixPlantae> <SuffixAlgae>-</SuffixAlgae> <SuffixFungi>-</SuffixFungi> <SuffixAnimalia>-</SuffixAnimalia> <BioCodeStatus>-</BioCodeStatus> <BacteriaStatus>-</BacteriaStatus> <BotanyStatus>-</BotanyStatus> <ZoologyStatus>-</ZoologyStatus> </Specification> </xs:appinfo> </xs:annotation> </xs:enumeration> <xs:enumeration value="taxinfragen"> <xs:annotation> <xs:documentation>[tax. infragen.] -- infrageneric tax. of undefined rank -- A name that appear between a genus name and a species epitheton and is not clearly marked as series or section, or other may be assigned to this rank until the true rank can be assigned by a taxonomic expert.</xs:documentation> <xs:appinfo> <Specification> <PreferredAbbreviation>tax. infragen.</PreferredAbbreviation> <AdditionalAbbreviations>-</AdditionalAbbreviations> <SuffixBacteriae>-</SuffixBacteriae> <SuffixPlantae>-</SuffixPlantae> <SuffixAlgae>-</SuffixAlgae> <SuffixFungi>-</SuffixFungi> <SuffixAnimalia>-</SuffixAnimalia> <BioCodeStatus>-</BioCodeStatus> <BacteriaStatus>-</BacteriaStatus> <BotanyStatus>-</BotanyStatus> <ZoologyStatus>-</ZoologyStatus> </Specification> </xs:appinfo> </xs:annotation> </xs:enumeration> <xs:enumeration value="subser"> <xs:annotation> <xs:documentation>[subser.] -- subseries</xs:documentation> <xs:appinfo> <Specification> <PreferredAbbreviation>subser.</PreferredAbbreviation> <AdditionalAbbreviations>*</AdditionalAbbreviations> <SuffixBacteriae>-</SuffixBacteriae> <SuffixPlantae>-</SuffixPlantae> <SuffixAlgae>-</SuffixAlgae> <SuffixFungi>-</SuffixFungi> <SuffixAnimalia>-</SuffixAnimalia> <BioCodeStatus>additional</BioCodeStatus> <BacteriaStatus>-</BacteriaStatus> <BotanyStatus>additional</BotanyStatus> <ZoologyStatus>-</ZoologyStatus> </Specification> </xs:appinfo> </xs:annotation> </xs:enumeration> <xs:enumeration value="ser"> <xs:annotation> <xs:documentation>[ser.] -- series</xs:documentation> <xs:appinfo> <Specification> <PreferredAbbreviation>ser.</PreferredAbbreviation> <AdditionalAbbreviations>*</AdditionalAbbreviations> <SuffixBacteriae>-</SuffixBacteriae> <SuffixPlantae>-</SuffixPlantae> <SuffixAlgae>-</SuffixAlgae> <SuffixFungi>-</SuffixFungi> <SuffixAnimalia>-</SuffixAnimalia> <BioCodeStatus>secondary</BioCodeStatus> <BacteriaStatus>-</BacteriaStatus> <BotanyStatus>secondary</BotanyStatus> <ZoologyStatus>-</ZoologyStatus> </Specification> </xs:appinfo> </xs:annotation> </xs:enumeration> <xs:enumeration value="subsect"> <xs:annotation> <xs:documentation>[subsect.] -- subsection</xs:documentation> <xs:appinfo> <Specification> <PreferredAbbreviation>subsect.</PreferredAbbreviation> <AdditionalAbbreviations>*</AdditionalAbbreviations> <SuffixBacteriae>-</SuffixBacteriae> <SuffixPlantae>-</SuffixPlantae> <SuffixAlgae>-</SuffixAlgae> <SuffixFungi>-</SuffixFungi> <SuffixAnimalia>-</SuffixAnimalia> <BioCodeStatus>additional</BioCodeStatus> <BacteriaStatus>-</BacteriaStatus> <BotanyStatus>additional</BotanyStatus> <ZoologyStatus>-</ZoologyStatus> </Specification> </xs:appinfo> </xs:annotation> </xs:enumeration> <xs:enumeration value="sect"> <xs:annotation> <xs:documentation>[sect.] -- section</xs:documentation> <xs:appinfo> <Specification> <PreferredAbbreviation>sect.</PreferredAbbreviation> <AdditionalAbbreviations>*</AdditionalAbbreviations> <SuffixBacteriae>-</SuffixBacteriae> <SuffixPlantae>-</SuffixPlantae> <SuffixAlgae>-</SuffixAlgae> <SuffixFungi>-</SuffixFungi> <SuffixAnimalia>-</SuffixAnimalia> <BioCodeStatus>secondary</BioCodeStatus> <BacteriaStatus>-</BacteriaStatus> <BotanyStatus>secondary</BotanyStatus> <ZoologyStatus>-</ZoologyStatus> </Specification> </xs:appinfo> </xs:annotation> </xs:enumeration> </xs:restriction> </xs:simpleType> |
simpleType TaxonomicRankSpeciesGroupEnum
| namespace | http://www.tdwg.org/schemas/tcs/1.01 | ||||||||||||||||||||||||
| type | restriction of xs:Name | ||||||||||||||||||||||||
| properties |
|
||||||||||||||||||||||||
| used by |
|
||||||||||||||||||||||||
| facets |
|
||||||||||||||||||||||||
| annotation |
|
||||||||||||||||||||||||
| source | <xs:simpleType name="TaxonomicRankSpeciesGroupEnum"> <xs:annotation> <xs:documentation>Subset of ranks; equivalent to BioCode "species group", i.e. only species and subspecies</xs:documentation> </xs:annotation> <xs:restriction base="xs:Name"> <xs:enumeration value="subsp_aggr"> <xs:annotation> <xs:documentation>[aggr.] -- subspecific aggregate (= group, complex) -- A loosely defined group of subspecies. Zoology: "Aggregate - a group of subspecies within a species. An aggregate may be denoted by a group name interpolated in parentheses."</xs:documentation> <xs:appinfo> <Specification> <PreferredAbbreviation>aggr.</PreferredAbbreviation> <AdditionalAbbreviations/> <SuffixBacteriae>-</SuffixBacteriae> <SuffixPlantae>-</SuffixPlantae> <SuffixAlgae>-</SuffixAlgae> <SuffixFungi>-</SuffixFungi> <SuffixAnimalia>-</SuffixAnimalia> <BioCodeStatus>-</BioCodeStatus> <BacteriaStatus>-</BacteriaStatus> <BotanyStatus>-</BotanyStatus> <ZoologyStatus>-</ZoologyStatus> </Specification> </xs:appinfo> </xs:annotation> </xs:enumeration> <xs:enumeration value="ssp"> <xs:annotation> <xs:documentation>[ssp.] -- subspecies -- Examples: Pinus nigra subsp. nigra Homo sapiens sapiens</xs:documentation> <xs:appinfo> <Specification> <PreferredAbbreviation>ssp.</PreferredAbbreviation> <AdditionalAbbreviations>subsp.; subspec.</AdditionalAbbreviations> <SuffixBacteriae>-</SuffixBacteriae> <SuffixPlantae>-</SuffixPlantae> <SuffixAlgae>-</SuffixAlgae> <SuffixFungi>-</SuffixFungi> <SuffixAnimalia>-</SuffixAnimalia> <BioCodeStatus>secondary</BioCodeStatus> <BacteriaStatus>covered</BacteriaStatus> <BotanyStatus>additional</BotanyStatus> <ZoologyStatus>additional</ZoologyStatus> </Specification> </xs:appinfo> </xs:annotation> </xs:enumeration> <xs:enumeration value="sp"> <xs:annotation> <xs:documentation>[sp.] -- species -- Examples: Taxus baccata, Homo sapiens</xs:documentation> <xs:appinfo> <Specification> <PreferredAbbreviation>sp.</PreferredAbbreviation> <AdditionalAbbreviations>spec.</AdditionalAbbreviations> <SuffixBacteriae>-</SuffixBacteriae> <SuffixPlantae>-</SuffixPlantae> <SuffixAlgae>-</SuffixAlgae> <SuffixFungi>-</SuffixFungi> <SuffixAnimalia>-</SuffixAnimalia> <BioCodeStatus>primary</BioCodeStatus> <BacteriaStatus>covered</BacteriaStatus> <BotanyStatus>principal</BotanyStatus> <ZoologyStatus>principal</ZoologyStatus> </Specification> </xs:appinfo> </xs:annotation> </xs:enumeration> </xs:restriction> </xs:simpleType> |
XML Schema documentation generated by XMLSpy Schema Editor http://www.altova.com/xmlspy